Clostridium thermocellum, a Gram-positive, thermophilic anaerobic bacterium, exhibits an outstanding capability for degrading cellulolytic biomass to release fermentable sugars of different lengths by means of its powerful cellulosomes.
- cellulase booster
- artificial cellulosomes
- designer cellulosomes
- laccases
- LPMOs
1. Introduction
During evolution, cellulolytic microbes developed an extracellular multienzyme complex called the cellulosomes to boost the cellulose degradation rate at maximum levels [1]. Since the polysaccharide compositions in plant cell walls vary substantially in both quality and quantity [2], C. thermocellum needs to mediate the composition of saccharolytic enzymes in the cellulosome complex to cope with the complexity and recalcitrance of a specific cell wall [3]. More than 70 cellulolytic enzymes-borne type-I dockerin (DocI) are noncovalently assembled onto the primary non-catalytic protein, termed scaffoldin (Scaf) CipA, through a calcium-dependent high-affinity interaction (i.e., dissociation constant K D < 10 −11 M) with the nine type-I cohesin (CohI) domains located on the CipA structure [4]. In addition, the CipA protein also contains a type-II dockerin (DocII) domain at its C-terminal to mediate the attachment of the cellulosome complex to the bacterial cell surface [5]. Correspondingly, C. thermocellum possesses three types of surface-anchoring Scaf(s), namely SdbA, Orf2p, OlpB, which contain one, two, and seven type-II cohesins (CohII), respectively, responsible for the binding of the cellulosome complex to the cell surface through CohII–DocII interaction.
Inspired by the Lego-like architecture of the C. thermocellum cellulosome, various research groups have been seeking to construct designer cellulosomes (DCs) for basic and applied studies [6][7][8]. Although there are several conjugation techniques to design artificial cellulosomes [9], the present review mainly focuses on the CohI– DocI interaction-based artificial cellulosome construction. Furthermore, the related issues of this approach, such as enzymatic unit positions, types of enzymes, linkers between DocI and catalytic domain, and number of carbohydrate-binding modules (CBMs), are updated and discussed. Another strategy is to recombinantly produce a library of individual catalytic enzymes with diverse hydrolytic reaction modes and then formulate the specified enzymatic cocktails for specific substrates [10]. In addition, to convert non-cellulolytic biofuels-producing microbes into consolidated bioprocessing (CBP)-enabling microbes, which can conduct enzyme production, substrate saccharification, and fermentation of the released sugars into biofuels in a single step, various studies have been carried out to express heterologous cellulolytic enzymes in heterologous host cells to make them genetically engineered cellulolytic microbes [11]. These engineered microbes use a cell-surface display or secretion system to display their novel hydrolytic capability. These strategies, with strengths and weaknesses, are discussed in the present review.
2. Conversion of Non-Cellulolytic Biofuel Microbes into Consolidated Bioprocessing Microbes
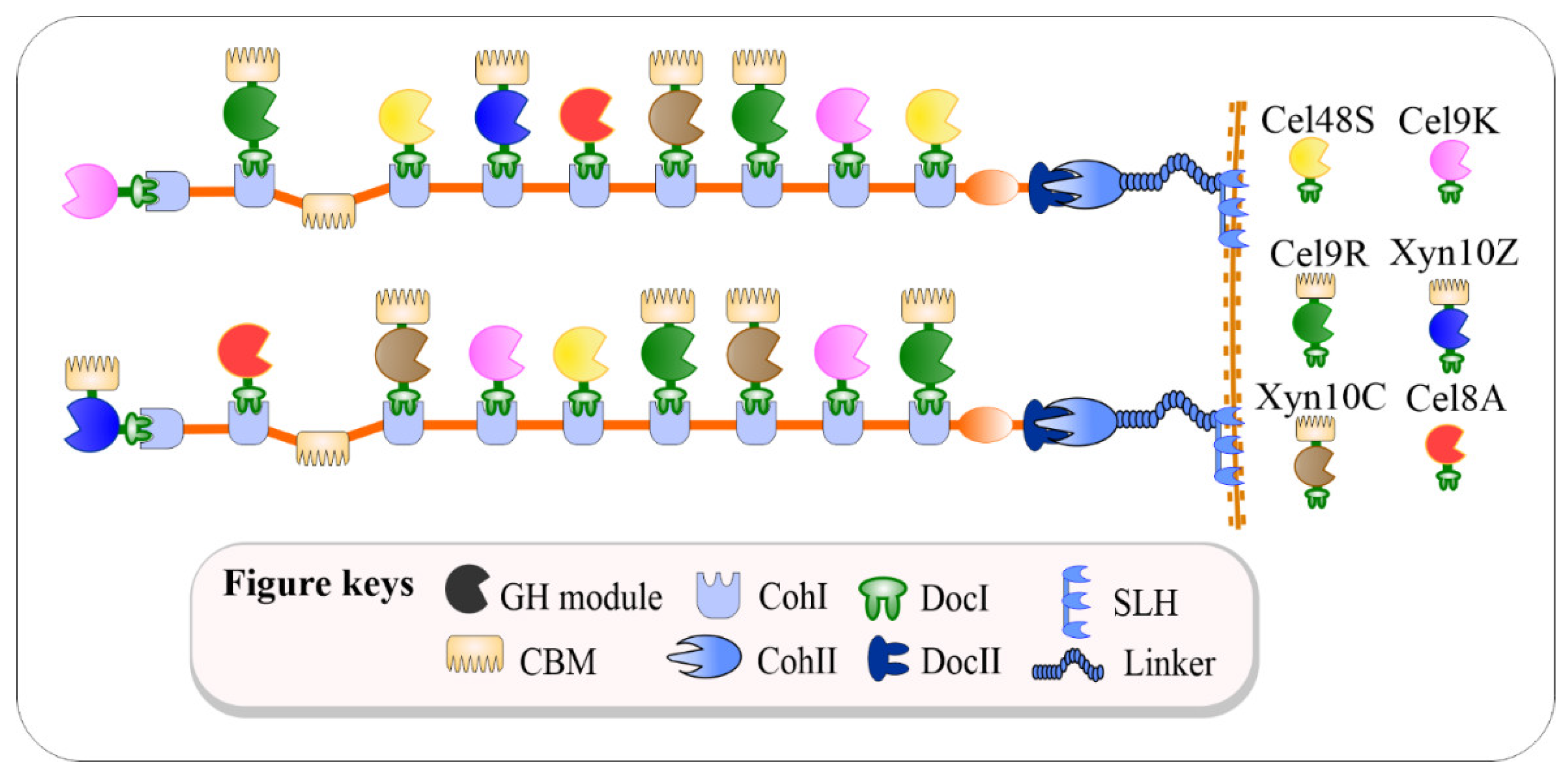
2.1. Creation of Cellulolytic Bacillus subtilis
2.2. CBP-Enabling Saccharomyces cerevisiae
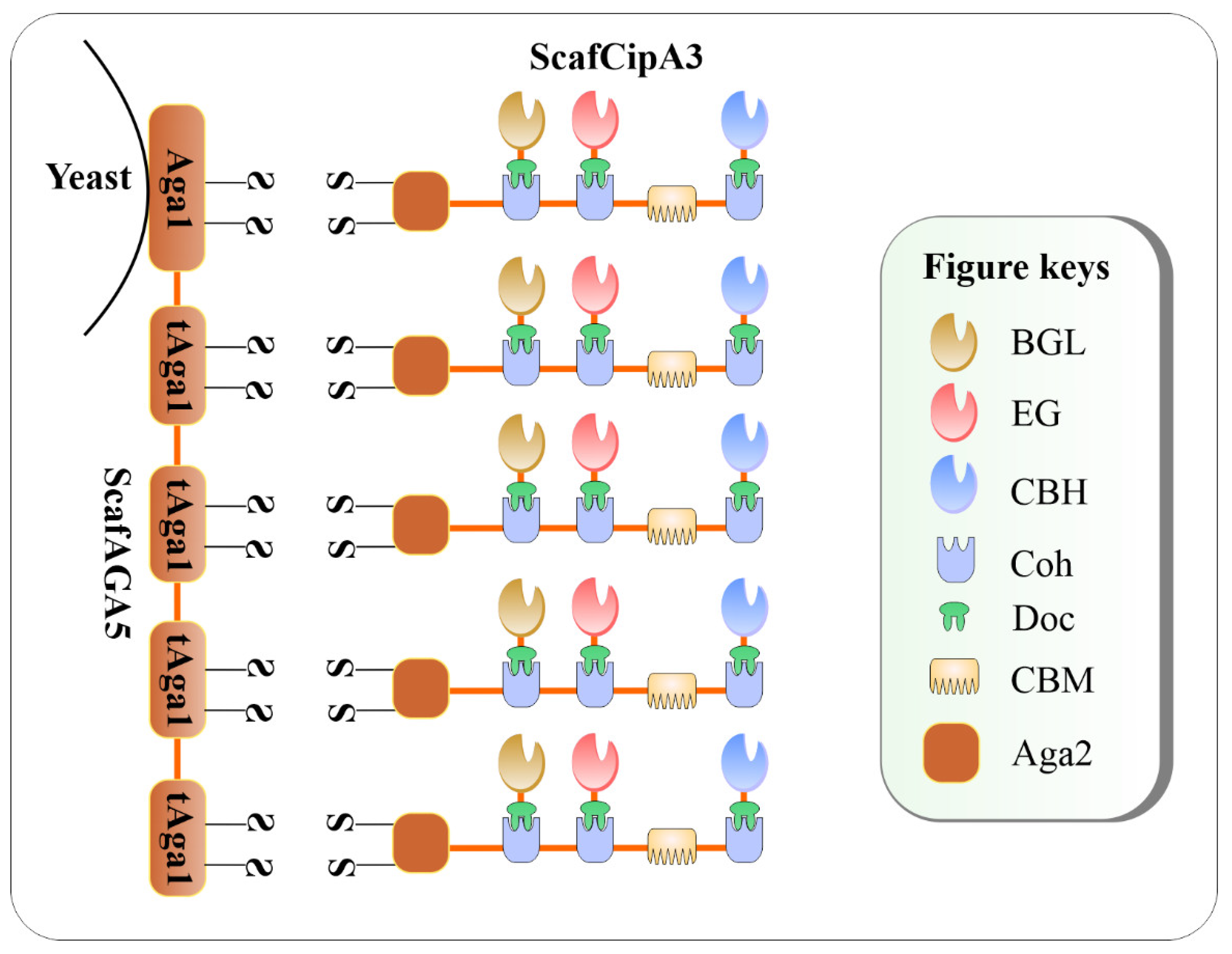
A double-layered cellulosome was synthesized in a study by Tang et al. [18] where an artificial ScafAGA3 bearing the repeated N-terminus of Aga1p (tAga1p) was displayed on the cell surface of S. cerevisiae through the Aga1p C-terminal domain. The ScafAGA3 was used as an anchoring protein via Aga1p-Aga2p linkage and the ScafCipA3 functioned as the primary Scaf for cellulase assembly (Figure 2). The ScafCipA3, which contained three CohI(s) from C. thermocellum on its structure, carried three cellulases from divergent microbes via CohI– DocI interaction. For the conversion of free cellulases into cellulosomal mode, a BGL from Saccharomycopsis fibuligera, a CBH from Talaromyces emersonii, and an EG CelA from C. thermocellum were fused with the traditional C. thermocellum DocI(s). The novel disulfide bonds showed higher display efficiency of the ScafAGA3 on the cell surface in comparison with that of the conventional CtScafCipA3. The result demonstrates that the covalent disulfide bonds of tAga1p–Aga2 appeared to outperform the non-covalent bonds of the conventional CohI–DocI pair in cellulase assemblage. For enzyme activity measurement, the enzymes were mixed with 5 mM p-nitrophenyl-β-D-cellobiose (pNPC) or carboxymethylcellulose sodium salt (CMC-Na) as the substrates in 50 mM citrate buffer (pH 5.0) at 50 °C for 30 min for CBH and EG measurements, respectively. The resultant engineered S. cerevisiae carrying the double-layered cellulosome ScafAGA5–ScafCipA3: CBH1/CelA/BGLA produced 1.52 g/L ethanol on 1% ( w / v ) PASC. In summary, despite many efforts to create the CBP S. cerevisiae , the bioethanol produced by these engineered strains have been still modest due mainly to the lower efficiency of the surface-displayed cellulosomes. Some catalytic proteins assembled onto the DCs did not have sufficient ability to digest high substrate concentrations to supply an abundant fermentable sugar source for the CBP S. cerevisiae.
2.3. CBP-Enabling Pichia pastoris
Pichia pastoris has been widely used for the expression of diverse recombinant proteins. The limited production of endogenous secretory proteins in P. pastoris makes the purification of recombinant proteins easy [19]. Additionally, an appropriate posttranslational modification is an advantage in producing proteins with correct folding and proper biological activity. Recently, an indirect P. pastoris surface-display method was developed to create a CBP cell factory [20]. Instead of using the usual non-covalent interactions between CohI and DocI from C. thermocellum, the colicin E7 DNase (CE7) and its matching immunity protein 7 (Im7) was used as a CE7–Im7 protein pair. The CE7 was mutated to inactivate DNase activity but retained its full binding affinity to generate CL7 tag, and the Im7, a cognate inhibitor of CE7, was employed to form an ultra-high-affinity IM7–CL7 protein pair (K D ~10 −14 –10 −17 M) [21]. The IM7–CL7 protein pair was used as an alternative to the conventional Coh-Doc pair for cellulosome assembly (Figure 3). A CBH from Yarrowia lipolytica, an EG Cel9D from C. thermocellum DSM1237, a BGL from Thermoanaerobacterium thermosaccharolyticum, and a CBM from T. fusca were fused with N-terminal CL7 tags and recombinantly expressed in E. coli. The surface anchoring protein SED from S. cerevisiae was fused to the IM7 scaffoldin protein to mediate the attachment of the enzyme complex onto cell surface. In turn, the IM7 proteins were engineered to display for two or three times to carry two to three catalytic modules, thus generating Y-IM2 and Y-IM3 yeasts, respectively. Subsequently, the engineered P. pastoris yeasts were incubated with the E. coli lysates containing cellulosomal-mode-cellulases and CBMs to promote the assembly of minicellulosomes on cell surface via IM7–CL7 interaction. On Avicel and PASC, the engineered Y-IM2 performed better than Y-IM3 with 2.5 g/L and 1.2 g/L ethanol, respectively, whereas the Y-IM3 surpassed Y-IM2 on CMC substrate with up to 5.1 g/L ethanol. Moreover, a synergistic effect on CMC hydrolysis was also observed in Y-IM3, indicating a positive relationship between the number of Scaf IM7 and the catalytic efficiency.
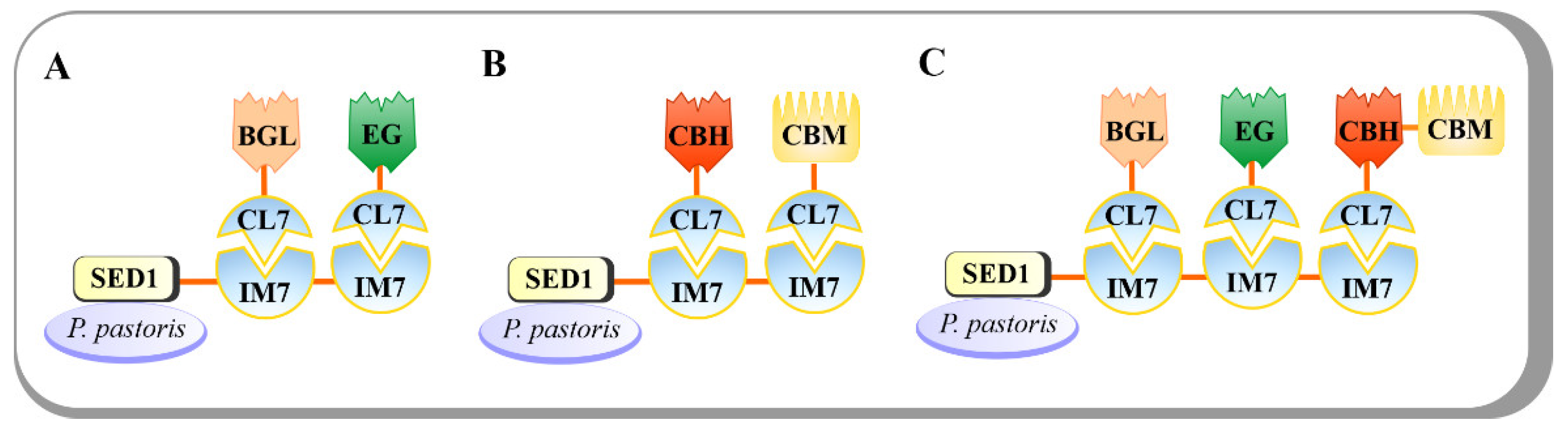
Figure 3. The novel ultra-high-affinity IM7–CL7 protein pair (KD~10−14–10−17 M) was used for cellulase assembly onto P. pastoris cell surface. (A) The engineered P. pastoris with one BGL and one EG enzyme on its surface. (B) The engineered P. pastoris with one CBH and one CBM on its surface. (C) The engineered P. pastoris with one CBH, one EG, one CBH, and one CBM on its surface.
2.4. Consolidated Bioprocessing-Enabling K. marxianus
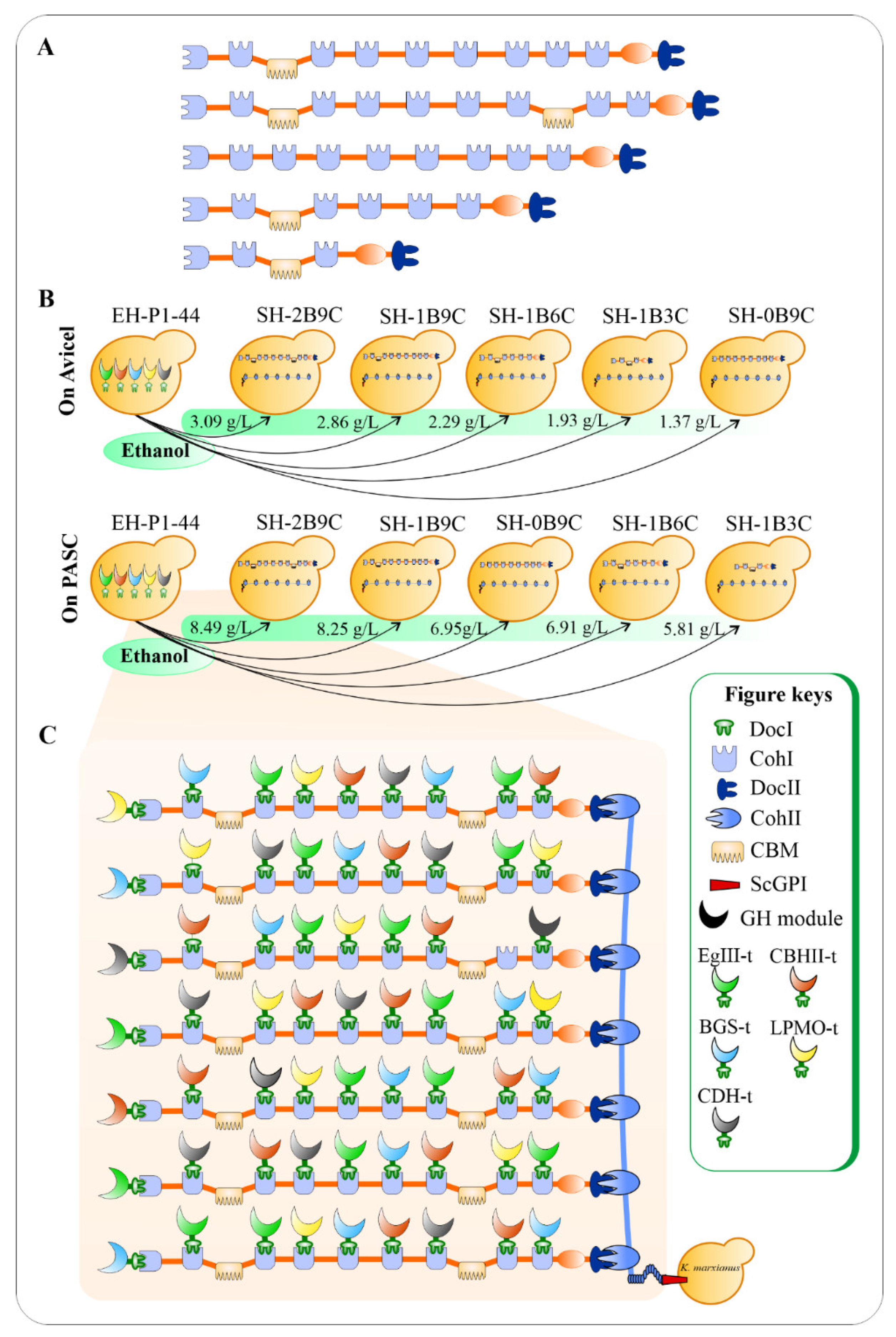
Apart from the LPMO from Thermoascus aurantiacus (TaLPMO) and its electron donor cellobiose dehydrogenase from Myceliophthora thermophila (MtCDH), an EG from T. reesei (TrEgIII), a synthetic CBH (CBHII), and a BGL from Neocallimastix patriciarum (NpaBGS) were fused with a DocI of C. thermocellum to create DocI-fused enzyme subunits, namely TrEgIII–t, CBHII–t, NpaBGS–t, TaLPMO–t, and MtCDH –t, respectively (Figure 4). In addition, a cell-surface protein glycosylphosphatidylonositol (GPI) from S. cerevisiae (ScGPI) was used to facilitate the anchoring of the artificial cellulosome to the K. marxianus cell surface because the anchoring domain of C. thermocellum is not suitable for eukaryotic hosts. The supernatants of red fluorescent protein (RFP)-fused DocT (RFP–DocT), OlpB–ScGPI, and CipA1B9C were mixed to form the entire cellulosome complex, namely OlpB– ScGPI :CipA1B9C:RFP–DocT, and their assembly was confirmed using an epifluorescence microscopy. The hydrolytic activity of cellulosome on 1% (w / v) Avicel was quantified by mixing the concentrated supernatants of the YP culture broth with Avicel and incubated at 40 °C for 6 h. On the model micro-crystalline cellulose Avicel, the amounts of sugar release were significantly influenced by the number of CohI and the number of CBMs, whereas the effect of these components was less pronounced on amorphous cellulose phosphoric acid swollen cellulose (PASC) hydrolysis (Figure 4 A,B). The results confirmed the decisive roles of the scaffoldin protein CipA in cellulose solubilization rate as elucidated in a previous study by Olson et al. [22]. To the best of our knowledge, this is the first time an assembly of 63 saccharolytic cellulosomal enzymes from different species was engineered and expressed on a heterologous host’s surface, enabling a greater biomass degradation rate for future consolidated bioprocessing (CBP) microorganisms ( Figure 4 C).
3. Promise, Limitations and Future Directions
This entry is adapted from the peer-reviewed paper 10.3390/catal11080996
References
- Zhivin-Nissan, O.; Dassa, B.; Morag, E.; Kupervaser, M.; Levin, Y.; Bayer, E. Unraveling Essential Cellulosomal Components of the (Pseudo)Bacteroides Cellulosolvens Reveals an Extensive Reservoir of Novel Catalytic Enzymes. Biotechnol. Biofuels 2019, 12, 1–21.
- Pauly, M.; Keegstra, K. Cell-wall Carbohydrates and Their Modification as a Resource for Biofuels. Plant J. 2008, 54, 559–568.
- Raman, B.; Pan, C.; Hurst, G.; Rodriguez, M., Jr.; McKeown, C.; Lankford, P.; Samatova, N.; Mielenz, J. Impact of Pretreated Switchgrass and Biomass Carbohydrates on Clostridium Thermocellum ATCC 27405 Cellulosome Composition: A Quantitative Proteomic Analysis. PLoS ONE 2009, 4, e5271.
- Mechaly, A.; Fierobe, H.-P.; Belaich, A.; Belaich, J.-P.; Lamed, R.; Shoham, Y.; Bayer, E. Cohesin-Dockerin Interaction in Cellulosome Assembly-A Single Hydroxyl Group of a Dockerin Domain Distinguishes Between Nonrecognition and High Affinity Recognition. J. Biol. Chem. 2001, 276, 9883–9888.
- Bayer, E.; Lamed, R.; White, B.; Flint, H. From Cellulosomes to Cellulosomics. Chem. Rec. 2008, 8, 364–377.
- Fierobe, H.-P.; Mingardon, F.; Mechaly, A.; Bélaïch, A.; Rincon, M.; Pagès, S.; Lamed, R.; Tardif, C.; Bélaïch, J.-P.; Bayer, E. Action of Designer Cellulosomes on Homogeneous Versus Complex Substrates. J. Biol. Chem. 2005, 280, 16325–16334.
- Moraïs, S.; Morag, E.; Barak, Y.; Goldman, D.; Hadar, Y.; Lamed, R.; Shoham, Y.; Wilson, D.; Bayer, E. Deconstruction of Lignocellulose into Soluble Sugars by Native and Designer Cellulosomes. mBio 2012, 3, e00508-12.
- Davidi, L.; Moraïs, S.; Artzi, L.; Knop, D.; Hadar, Y.; Arfi, Y.; Bayer, E. Toward Combined Delignification and Saccharification of Wheat Straw by a Laccase-Containing Designer Cellulosome. Proc. Natl. Acad. Sci. USA 2016, 113, 10854–10859.
- Gunnoo, M.; Cazade, P.-A.; Galera-Prat, A.; Nash, M.; Czjzek, M.; Cieplak, M.; Alvarez, B.; Aguilar, M.; Karpol, A.; Gaub, H.; et al. Nanoscale Engineering of Designer Cellulosomes. Adv. Mater. 2016, 28, 5619–5647.
- Leis, B.; Held, C.; AndreeBen, B.; Liebl, W.; Graubner, S.; Schulte, L.-P.; Schwarz, W.; Zverlov, V. Optimizing the Composition of a Synthetic Cellulosome Complex for the Hydrolysis of Softwood Pulp: Identification of the Enzymatic Core Functions and Biochemical Complex Characterization. Biotechnol. Biofuels 2018, 11, 1–15.
- Fan, L.; Zhang, Z.; Yu, X.; Xue, Y.; Tan, T. Self-Surface Assembly of Cellulosomes with Two Miniscaffoldins on Saccharomyces Cerevisiae for Cellulosic Ethanol Production. Proc. Natl. Acad. Sci. USA 2012, 109, 13260–13265.
- Lambertz, C.; Garvey, M.; Klinger, J.; Heesel, D.; Klose, H.; Fischer, R.; Commandeur, U. Challenges and Advances in the Heterologous Expression of Cellulolytic Enzymes: A Review. Biotechnol. Biofuels 2014, 7, 1–15.
- Cui, W.; Han, L.; Suo, F.; Liu, Z.; Zhou, L.; Zhou, Z. Exploitation of Bacillus Subtilis as a Robust Workhorse for Production of Heterologous Proteins and Beyond. World J. Microbiol. Biotechnol. 2018, 34, 1–19.
- Chang, J.; Anandharaj, M.; Ho, C.; Tsuge, K.; Tsai, T.; Ke, H.; Lin, Y.; Ha-Tran, D.; Li, W.; Huang, C. Biomimetic Strategy for Constructing Clostridium Thermocellum Cellulosomal Operons in Bacillus Subtilis. Biotechnol. Biofuels 2018, 11, 1–13.
- Nishizaki, T.; Tsuge, K.; Itaya, M.; Doi, N.; Yanagawa, H. Metabolic Engineering of Carotenoid Biosynthesis in Escherichia Coli by Ordered Gene Assembly in Bacillus Subtilis. Appl. Environ. Microbiol. 2007, 73, 1355–1361.
- Tsuge, K.; Matsui, K.; Itaya, M. One Step Assembly of Multiple DNA Fragments with a Designed Order and Orientation in Bacillus Subtilis Plasmid. Nucleic Acids Res. 2003, 31, e133.
- Stern, J.; Kahn, A.; Vazana, Y.; Shamshoum, M.; Moraïs, S.; Lamed, R.; Bayer, E. Significance of Relative Position of Cellulases in Designer Cellulosomes for Optimized Cellulolysis. PLoS ONE 2015, 10, e0127326.
- Tang, H.; Wang, J.; Wang, S.; Shen, Y.; Petranovic, D.; Hou, J.; Bao, X. Efficient Yeast Surface-Display of Novel Complex Synthetic Cellulosomes. Microb. Cell Factories 2018, 17, 1–13.
- Karbalaei, M.; Rezaee, S.; Farsiani, H. Pichia Pastoris: A Highly Successful Expression System for Optimal Synthesis of Heterologous Proteins. J. Cell. Physiol. 2020, 235, 5867–5881.
- Dong, C.; Wang, X.; Sun, W.; Chen, L.; Li, S.; Wu, K.; Ma, L.; Liu, Y. Engineering Pichia Pastoris with Surface-Display Minicellulosomes for Carboxymethyl Cellulose Hydrolysis and Ethanol Production. Biotechnol. Biofuels 2020, 13, 1–9.
- Vassylyeva, M.; Klyuyev, S.; Vassylyev, A.; Wesson, H.; Zhang, Z.; Renfrow, M.; Wang, H.; Higgins, N.; Chow, L.; Vassylyev, D. Efficient, Ultra-High-Affinity Chromatography in a One-Step Purification of Complex Proteins. Proc. Natl. Acad. Sci. USA 2017, 114, 5138–5147.
- Olson, D.; Giannone, R.; Hettich, R.; Lynd, L. Role of the CipA Scaffoldin Protein in Cellulose Solubilization, as Determined by Targeted Gene Deletion and Complementation in Clostridium Thermocellum. J. Bacteriol. 2013, 195, 733–739.
