Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biology
β-glucans are complex polysaccharides that are found in several plants and foods, including mushrooms. β-glucans display an array of potentially therapeutic properties.
- β-glucan
- clinical trials
- biomedicine
- immunomodulation
- metabolism
1. Introduction
β-glucans/Beta-glucans are a large class of complex polysaccharides that can be found in an abundance of sources. Depending on origin, β-glucans can be classified as cereal or non-cereal derived. Cereal sources of β-glucans include oat and barley and non-cereal sources can include mushroom, algae, bacteria and seaweed [1]. β-glucans are biologically active compounds that have been widely reported to improve health [2]. Specific to this group of polysaccharides is a 1,3 beta-glycosidic linked backbone; separate to this, the polysaccharide can take many forms, dictated by origin.
There is a growing interest in foods that have the potential to lower the risk or incidences of chronic diseases or promote lifespan as well as have anti-aging properties. This has led to an increase in awareness of the effect of diet on health [3][4].
In 1979, Stephen DeFelice devised the term nutraceutical, which may be isolated nutrients, dietary supplements, genetically engineered foods and herbal products [5]. Nutraceuticals are defined as a food or food component that provides medical or health benefits, including prevention and/or treatment of disease [5]. Similarly, bioactive compounds are defined as “essential and non-essential compounds that occur in nature, are part of the food chain and are shown to have an effect on human health” [6].
Bioactive substances in food provide health benefits beyond the nutritional benefits of the product [7]. β-glucans are reported to be both a bioactive and a nutraceutical. Their therapeutic effects can also be largely classified into two categories, metabolic/GI effects or immune-modulatory effects, which is largely based on structure, determined by source [1][8].
Metabolic effects are usually observed with cereal derived β-glucans. Effects include modulation of the gut microbiome, cholesterol reduction and decreased cardiovascular and diabetic risk. Non-cereal β-glucans are associated with immune-modulatory effects, anti-tumor effects, wound healing and alleviation of immune-related conditions, as demonstrated in Figure 1 [1]. β-glucans are also administered as an animal and fish feed additive to increase [9][10]. These molecules also have applications in the food industry for thickening and for gelation purposes [11].
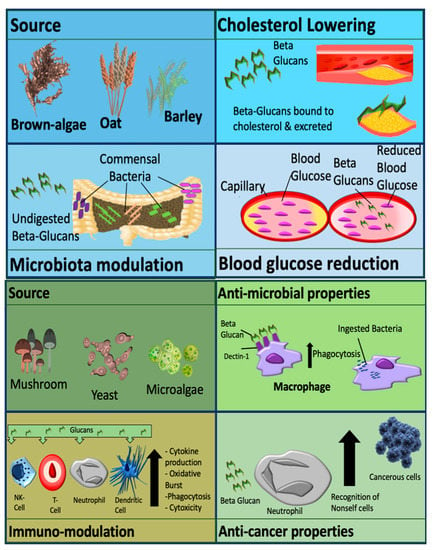
Figure 1. Mechanisms and activity of β-glucan which are dependent on source. β-glucan can be classified as cereal derived (upper panel), or non-cereal derived (lower panel). Picture originally published in [1]. Modified with permission.
In this encyclopedia entry on β-glucans, we start from their initial discovery, then examine β-glucan sources, characterize their complex and diverse structures, and examine the implications of their structural variations on their activity profile. The therapeutic potential in different disease conditions is then discussed, and the barriers to fully realizing this potential is dissected in some detail. Finally, we examine other uses of β-glucan in animal health and their application in the food industry.
2. History of β-Glucans
As β-glucans are a structural component of mushrooms, their use as an unknown therapeutic in medicinal mushrooms dates back hundreds of years [12]. β-glucans, as an active compound, followed two different pathways of discovery. In Western medicine, they were initially discovered by Pilliner et al., in 1954 [13]. β-glucans were discovered unintentionally through the discovery of Properdin in the compliment system. Zymosan was the stimulatory agent used in these experiments. It is a crude mixture of yeast cell wall particles, which is now established to contain high levels of β-glucans. In 1961, Riggi and Dilugio identified the active compound in zymosan to be a polysaccharide. They concluded that the polysaccharide required a 1,3 beta type linkage for activity [13].
Independently, concurrently in Japan, β-glucans were being investigated for antitumor properties, with the first study in 1969. Chikara et al. isolated β-glucans from the mushroom Lentinus edodes and demonstrated it to have an ability to inhibit sarcoma in mice [14]. β-glucans were previously used as a traditional medicine for cancer therapy [15][16][17]. To date, β-glucans isolated from Lentinus edodes named Lentinan and Polysaccharide K are two licensed drugs in Japan [18].
Developments
In the wake of these pioneering studies, β-glucan research has gained considerable attention and advancements, with these bioactives being registered in clinical trials for an array of conditions including inflammatory conditions, cardiometabolic diseases, obesity and cancer, and more recently, as a nutritional supplement for the treatment of COVID-19 in conjunction with vaccination (Efficacy and Tolerability of ABBC1 in Volunteers Receiving the Influenza or Covid-19 Vaccine—Full Text View—ClinicalTrials.gov) [19]. The US Food and Drug Administration has also recognized the benefits of consuming β-glucans, recommending a 3 g/day intake of oat bran, a source of β-glucan and registered as a cholesterol-reducing food [20].
3. Sources and Structure of β-Glucans
As mentioned, β-glucans can be classified by source: cereal or non-cereal. This classification is largely based on structure, as β-glucans from cereal have a different structure than those originating from non-cereal sources. Secondly, β-glucans from cereal sources can differ; for example, β-glucans from barley can be structurally different to those of oat. The same diversity can occur between β-glucans from non-cereal sources. All β-glucans are homo-polysaccharides and essentially composed of glucose units linked together and thus have a characteristic 1,3 linked backbone [21][22], which is fundamental to activity [23]. The structural difference occurs at branching off this backbone, which is dictated by source. β-glucans can be unbranched or branched [24]. Branching can usually occur at either the 1,4 or 1,6 position [24]. These molecular and structural characteristics will determine functional activity as β-glucans have a defined structure–activity relationship [25].
Cereal or grain derived β-glucans usually have 1,3 1,4 glycosidic linkages without any 1,6 bonds or branching [26][27]. Non-cereal sources usually have 1,6 linked branches off the main side chain. Figure 2 demonstrates the structural differences between β-glucans. Other glucans have no branching, such as Curdlan, a glucan isolated from Agrobacterium that contains no side branching, just a beta-D glucan backbone [28]. There are, however, exceptions—Sorghum arundinaceum, an ancient cereal grain, was found to contain β-glucans with alpha 1,4 linked D-glucopyranose residues with 1,3, 1,6 branching points [29]. Moreover, different species of Sorghum have different structures; Sorghum bicolor contains 1,3 with 1,4 linkages [30].
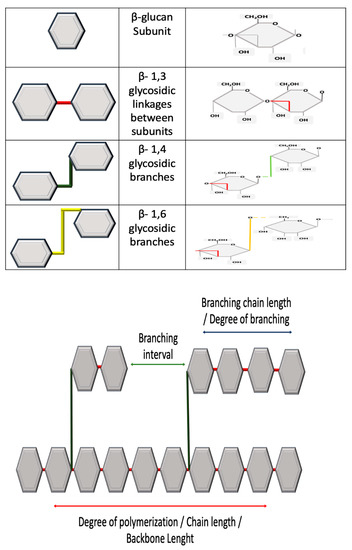
Figure 2. All β-glucans have a 1,3 linked backbone. The variances in structure occur with glycosidic side branches. Variances can also occur in chain length or degrees of polymerization, branching interval and branch length. Source and extraction technique will influence final structure. Various analytical methods can be used for the characterization of β-glucans, as outlined in Table 1.
β-glucans from the same species source may have variances in structure. Variances can include chain length, degrees of branching, polymerization and 3D conformational structure. Three-dimensional conformational structure can be random coil, single helix or triple helix [2]. Furthermore, other factors that can influence the structure include growth conditions, extraction procedure and analysis [20][31].
It has been shown that β-glucans specifically of mushroom origin have anti-tumor properties. Lentinan is a β-glucan isolated from mushroom which has a β-helix conformation [32][33]. If the helix is destroyed or removed, the anti-tumor activity decreases. Other studies demonstrated that molecular weight and chain conformation are all dependent on anti-tumor activity [34]. The ability of β-glucans to interact and modulate immunity is correlated to polymer length, degree of branching and tertiary structure [23][35]. Larger β-glucans have been shown to activate leucocytes directly. Activation initiates phagocytosis, antimicrobial activities and production of cytokines. Medium to smaller-sized β-glucans induce NF-κΒ and mediate inflammation [36].
β-glucans can also be isolated from macro and microalgae sources. Microalgae are unicellular photosynthetic organisms, whereas macroalgae are larger multicellular organisms. Laminarian is a water-soluble β-glucan extracted from the cell wall storage of Eisenia bicyclis, a brown macroalgae. Similarly, to other non-cereal sources, the structure consists of a 1,3 linked backbone with 1,6 side branching. The glycosidic bonds have different ratios depending on the habit or season of extraction [37][38].
Laminarian possesses anti-inflammatory, anti-apoptotic, anti-tumor, antioxidant and anticoagulant properties. More recently, anti-cancer effects via enhanced apoptotic cellular death and angiogenic potential have been reported [37]. This polysaccharide is also being exploited as a carrier for gene delivery [39]. Topical laminarian-based creams improve wound healing via collagen deposition and promotion of skin regeneration [40]. Oral supplementation of laminarian has also been shown to suppress the progressive development of precancerous lesions in mice [38].
Soluble β-glucan from macro algae has been applied in the aquaculture industry to fish in the early stages of development as well as cultures of crustaceans as a bath treatment to enhance immunity in early life [41]. Species of brown algae (Phaeophyceae) and red algae (Gracilariaceae) contain a 1,3 1,6 linked β-glucan with immunomodulatory properties—via activation of spleen lymphocytes [41].
The microalga Euglena gracilis is a well reported source of β-glucans. The microalga is of the division Euglenoophyceae and family Euglenales [42][43]. β-glucans isolated from this source are called paramylon, which are large, linear, unbranched 1,3 β-glucans with the potential to support immune function [44]. A clinical trial that investigated the effects of supplementation with E. gracilis that contained a 50% β-1,3-glucan on immune function reduced the severity of upper respiratory tract infections (URTI), decreasing sick days and symptoms compared to placebo [43].
Extraction, Purification and Characterisation Methods
There are four types of extraction procedures used for isolation of β-glucans: alkaline extraction, acidic extraction, enzymatic extraction and water extraction. More advanced methods can include ultrasound-assisted extraction, microwave-assisted extraction and superheated water extraction.
Cereal derived β-glucans are generally located in the aleurone (proteins stored as granules), in the sub-aleurone or in the cell wall of endospores, which are all located in oat, barley, wheat and rice [45]. Extractions from cereal sources can be difficult and therefore, β-glucans from these sources are commercially more expensive [46]. There is no standard method for the extraction of β-glucans. Most researchers modify commonly published methods. The method selected is usually based on solubility of the β-glucan in hot water or solvents and is followed by precipitation using 2-propanol or ethanol [47]. Other published methods include accelerated solvent extraction (ASE) [48], sodium hydroxide as initial extraction solvent [47]. Table 1 lists the various extraction methods for β-glucans as well as analytical methods for identification.
For β-glucans from non-cereal sources, hot water extraction is the most common method [49]. Samples are usually milled, extracted with water and then precipitated with solvents such as ethanol [50]. Other methods have used ethanol precipitation and chromatography methods using Sephadex or Di-Ethyl-Amino-Ethyl (DEAE) [47][51].
Simple extraction techniques will isolate the β-glucans to some extent. Usually, there are other contaminating materials present, e.g., proteins. Enzymatic techniques can be used for the removal of proteins. The isolated β-glucans, by simple extraction techniques, are usually fit for purpose. Purification and extensive extraction techniques can be expensive and tedious. However, different extraction and purification techniques will yield different products and cause inconsistences between results. A way to control this is to fully characterize β-glucans after extraction, as it is important that β-glucans retain their structural characteristics after extraction [52]. Methods applied for characterization include gel permeation chromatography (GPC) [53] for molecular weight estimation, Fourier-transform infrared spectroscopy (FTIR) and nuclear magnetic resonance spectroscopy (NMR) for structural characterization [54][55]. For the detection of β-glucans in products, enzymatic reactions are also available [56].
Table 1. Extraction and characterization methods for β-glucan.
| Source | Extraction Method | Analytical Method | Ref |
|---|---|---|---|
| Hull-less Barley Bran | Ultrasonic extraction, Hot water extraction, Microwave extraction, Microwave assisted ultrasonic extraction |
Megazyme commercial quantification kit, Structure determined using FTIR, Molecular weights determined using gel permeation chromatography (GPC) |
[57] |
| Hull-less Barley | Alkali extraction and ethanol precipitation. | High pressure size exclusion chromatography (HPSEC), Methylation, Gas Chromatography Mass Spectroscopy (GC-MS) |
[21] |
| Barley | Ethanol Extraction and enzyme treatment with amylase. | Megazyme commercial quantification kit, Molecular weight was determined using size-exclusion ultra-high-performance liquid chromatography |
[58] |
| Barley Bran | Enzyme extraction using α-amylase, protease, glucoamylase, pullunlanase and xylanase | Megazyme commercial quantification kit, HPSEC, Rheology, SEM |
[59] |
| Barley | Pressurized aqueous ethanol | Megazyme commercial quantification kit, GPC |
[60] |
| Barley | Ethanol and water extraction | N/A | [61] |
| Oat and Barley | Multistage approach; Solvent (acetone) and enzyme extraction α-amylase and hot water extraction | Megazyme commercial quantification kit, Asymmetric flow field-flow fractionation (AF4) coupled to multiangle light scattering (MALS), differential refractive index (dRI) and fluorescence (FL) detection, High performance anion exchange chromatography |
[62] |
| Oat | Multistage enzymatic and solvent extraction—enzymes α-amylase, amyloglucosidase, and papain | Megazyme commercial quantification kit | [63] |
| Oat | Subcritical-water extraction | Megazyme commercial quantification kit HPLC |
[64] |
| Oat | Enzymatic extraction- α-amylase | Megazyme commercial quantification kit | [65] |
| Corn pericarp | Anion exchange chromatography and affinity chromatography | HPLC, NMR, Methylation |
[66] |
| Castanea mollissima | Water extraction followed by ethanol extraction purified using anion exchange chromatography | Phenol- sulfuric acid method, High performance gel permeation chromatography (HPGPC) HPLC, FTIR, Methylation analysis |
[67] |
| Pueraria lobata | Ethanol extraction followed by cold water extraction followed by further ethanol extraction. Fractions of extracts collected using DEAE-Sepharose chromatography column | Carbohydrate content determined by phenol-sulfuric acid method, HPGPC, Congo red method used to determine triple helix structure. |
[68] |
| Ziziphus jujuba Mill | 3 Phase extraction, Aqueous alkaline extraction; acidic precipitation of proteins at their isoelectric point, precipitation of glucans with absolute ethanol |
Megazyme commercial kit, FTIR, Surface structural differences determined using scanning electron microscope. |
[69] |
| Tuber melanosporum | Extracted pressurized liquids. Water and ethanol used as extraction solvents. | Analyzed by NMR and Gas chromatography mass spectroscopy GC_MS | [70] |
| Phaseolus vulgaris | Sonication of cell wall residue | Gel filtration chromatography, Methylation analysis, NMR |
[71] |
| Punica granatum | Alkaline treatment, isoelectric precipitation, alcohol precipitation | FTIR | [72] |
| Euglena cantabrica | Pressurized liquid extraction (PLE) using different temperature 40–180 °C using green solvents (ethanol-water) mixtures. | Extracts analyzed by high-pressure size-exclusion chromatography coupled to an evaporative light-scattering detector. | [73] |
| Laminaria hyperborea | Hydrothermal assisted extraction—acid extraction, temperature and pressure treatment, filtration and freeze drying. | Concentration calculated using Megazyme commercial kit. | [74] |
| Saccharomyces cerevisiae | Hot water Extraction & Enzymatic Treatment; enzymes-protease and lipase | Concentration calculated using Megazyme commercial kit. | [75] |
| Saccharomyces cerevisiae | n/a | Congo red assay—colorimetric | [76] |
| Saccharomyces cerevisiae | Yeast was cultured in yeast extract-peptone-glucose (YBG) broth to produce ß-glucans. Cells were sonicated. Alkaline-acid extraction used as extraction method. | n/a | [77] |
| Saccharomyces cerevisiae | Acid-base extraction method | FTIR analysis of structure, HPLC |
[78] |
| Saccharomyces cerevisiae | Cell exposure to hot water (autoclaving), thermally induced autolysis, homogenization in a bead mill, sonication | FTIR, Megazyme commercial kit |
[79] |
| Saccharomyces cerevisiae | Alkaline and acidic extraction | FTIR, NMR |
[80] |
| Pleurotus eryngii | Water extraction at different temperatures and pressures | High pressure size exclusion chromatography (HPSEC), Gel permeation chromatography (GPC) |
[81] |
| Cantharellus tubaeformis | Pressurized hot water extraction (80–240 °C) | Megazyme commercial kit | [82] |
| Pleurotus sajor caju | Hot aqueous extraction | NMR spectroscopy, HPSEC, Methylation analysis |
[83] |
| Agaricus bisporus, Lentinula edodes and Pleurotus ostreatus | Pressurized water extraction (PWE) | Megazyme commercial kit | [84] |
| Mushroom by-products | Mechanical agitation and ultrasound assistance in ethanol/water solutions | Megazyme commercial kit | [85] |
| Ganoderma lucidum | Hot water extraction, Soxhlet extraction, ultrasound assisted extraction. | Phenol Sulfuric acid assay, HPGPC, Content and ration of branching determined by enzymatic-HPAEC-PAD detection, FTIR, SEM |
[86] |
| Pholiota nameko | Defatting process with cold water, Hot aqueous extraction Enzyme treatment with amylase |
NMR, Methylation |
[87] |
| Pleurotus ostreatus | Methanol extraction. Hot water extraction; Acid hydrolysis |
Megazyme commercial kit, GPC, Hydrophilic interaction chromatography |
[88] |
| Lentinus edodes- | Orthogonal alkaline extraction with sodium hydroxide—further purification ethanol precipitation and anion exchange chromatography | 1H-NMR High-performance gel permeation chromatography–refractive index–multi-angle laser light scattering (HPGPC-RI-MALLS) Methylation analysis |
[89] |
4. Activity
4.1. Absorption
Although other routes of administration of β-glucans have been explored, oral administration is the most common, as β-glucans are a food-derived compound. When orally administered, studies have shown that small β-glucan concentrations in other tissues are low. Studies have also demonstrated that β-glucans reach the small intestine intact. The primary reaction occurs between the molecule and the gut epithelia [90]. Peyer’s patches are groupings of lymphoid follicles in the ileum region of the small intestine. It is in this location that immune stimulatory components are taken up to lymphatic circulation. In the Peyer’s patches, there are circulating antigen presenting cells (APCs) [91]. When β-glucans reach the Peyer’s patches, dendritic cells or microfold (M cells) recognize β-glucans and present them to other immune cells located here [92][93].
Pathogen-associated molecular patterns (PAMPs) are essentially the signature markings of pathogens. It is how they are recognized by immune counterparts. PAMPs that contain a sugar component such LPS from Gram-negative bacteria are classified as sugar complexed PAMPs (SCPs). SCPs are functional and structural components of the pathogen [94]. Receptors recognize these unfamiliar microbial structures and respond. β-glucans are a structural component of fungi and therefore are recognized as a PAMP. This is beneficial as β-glucans initiate an immune response without introducing an infectious agent such as a fungus or bacteria. As PAMPs, β-glucans can stimulate immune cells in isolated form—i.e., they are not required to be attached to the infectious agent—they have been developed as adjuvants and immunotherapeutics [95].
There is huge diversification of carbohydrate structure and branching, creating a range of complex monomers and polymers, and there are specific receptors to recognize these conformations which elicit the necessary response [96][97]. The PAMP recognition ability of the immune cells requires receptors, namely, pathogen recognition receptors (PRRs) [98].
4.2. Cellular Activation
It is well established that cellular biological activities—namely, immune modulatory activity—are dependent on recognition and binding of receptors. C-type lectin receptors (CLRs) are the main pathogen recognition receptors involved in fungal recognition [99]. Initial or broad-spectrum recognition of β-glucans is based on the 1,3 backbones usually by a receptor called Dectin-1 or the β-glucan receptor [92]. It is reported that β-glucans activate intestinal epithelial cells to secrete pro-inflammatory cytokines in a Dectin-1 and Syk-dependent manner [100].
Dectin-1, a CLR-type receptor, is present on macrophages, dendritic cells, neutrophils and on pathways where pathogens invade [101][102][103]. After activation, Dectin-1 will trigger the NF-κΒ pathway, which will induce cytokine synthesis by Syk-dependent pathways. Pathways can include NF-κΒ inducting kinase (SYK-NIK) pathways or Syk independent pathways, including the Ras associated factor-l (RAF-1) pathway [104].
Complement receptor 3 (CR3) is mainly expressed on NK cells, dendritic cells, macrophages and neutrophils [105]. This specific receptor usually responds to iC3B opsonized cells and pathogens [106].
CR3 is involved in the recognition of endogenous ligands, but also acts as an opsonic receptor for the complement component and as a non-opsonic receptor for β-glucan. The anti-tumorigenic properties of β-glucans are correlated to recognition of this receptor and deficiency is correlated to a loss in activity [107].
Other receptors that recognize β-glucan include lactosylceramide receptor, scavenger receptors and Toll like receptors (TLR), namely TLR2 [108][109]. TLR 2,4 and 6 co-bind to dectin-1 after glucan recognition [110]. Lactosylceramide receptors are found in plasma membrane of many different cells [95]. Binding and recognition of these receptors induces ingestion, respiratory burst, microbial killing, inflammatory processes and the release of chemical mediators that activate and recruit other cells [111].
Dectin-1 can interact with β-glucans over a wide range of affinities; although 1,6 branching does not influence binding, branched β-glucans appear to have a stronger affinity in comparison to linear β-glucans [112]. Other influences over recognition include chain conformation and polymerization. Extraction procedure can have a huge impact on β-glucan conformation [113]. Thus, the isolation and purification procedures must be carefully selected, as it will ultimately influence the activity of β-glucans.
5. Therapeutic Potential and Challenges
5.1. Therapeutic Potential
In May 2006, the US Food and Drug Administration acknowledged that β-glucans reduce the risk of coronary heart disease. Health claims relating to β-glucans specifically for maintenance of blood cholesterol were approved in Europe in 2009 [114].
From a metabolic viewpoint, β-glucans have the ability to lower cholesterol and improve glycemic control [115][116][117]. Viscous β-glucans have been shown to modulate host bile acid metabolism [117]. In lowering cholesterol and triglycerides, the risk of cardiovascular diseases is also reduced [118]. Yeast β-glucans serve as prebiotics, improving gastrointestinal function by enhancing intestinal microbiota [1][119].
Immune modulatory properties include stimulation of the immune response and initiation of inflammatory processes as well as improving resistance to infections [120]. Non cereal β-glucans alleviate allergic conditions [121][122]. β-glucans have also been shown to enhance bacterial clearance from blood and reduce mortality in a rat model of intra-abdominal sepsis [123].
Immuno-oncology is the study of treatments that utilize the immune system to target cancer [124]. β-glucans have shown promise in this area as they stimulate immune cells to target cancerous cells. β-glucans have been demonstrated to induce macrophages and NK cells to attack and destroy cancerous cells [125]. Mushroom β-glucans promoted T-cell immunomodulation and neutrophil infiltration into tumors in mice, which lead to tumor growth inhibition [126]. Oral administration of β-glucans has similar effects to other routes of administration when used as an anti-tumor immunotherapy [107].
Promising results in clinical trials using β-glucans as an adjuvant in conjunction with monoclonal antibodies for cell mediated tumor killing has established them as a potential intervention in the field of immune-oncology [127].
β-glucans have also gained attention as vaccine adjuvants. When combined with vaccines, they have the ability to enhance sensitivity by stimulating immune cells [128]. Stimulants that are used for immune modulatory functions are referred to as immune adjuvants [129]. As β-glucans are recognized by immune counterparts as PAMPs, they are widely investigated for their potential as immune adjuvants [130][131]. They have therefore been administered as adjuvants in vaccines or counterparts in vaccine delivery systems [132][133].
5.2. Therapeutic Challenges
The therapeutic promise of β-glucans for an array of clinical conditions is considerable [127]. The translation to clinical application is not as successful for several reasons. Firstly, like all naturally derived products, β-glucans can represent a complex mixture of ingredients which can all contribute to or prevent activity. Extraction methods will greatly affect the final compound. Intense extraction procedures can alter the natural configuration of the β-glucan compound. Softer extraction procedures can leave other contaminating residues present, which can dilute, contaminate or even contribute to activity [27].
Secondly, there is huge diversity among β-glucan structures. Diversity occurs between β-glucans from different species, but there are also variances in extracted samples from the same species. These differences include degree of branching, monosaccharide composition, linkage ratio and linkage type [134]. The molecular weight of the β-glucan greatly influences activity, which can be dictated by growth conditions and the extraction procedure. For example, β-glucans isolated from barley in one experiment was shown to have a molecular weight of 3 × 104 g/mol and in a separate experiment found to have a molecular weight of 270 × 104 g/mol [31]. The distribution of polysaccharides from aleurone to endosperm in cereal sources can also vary depending on origin. In one study, aleurone was found to contain 26% w/w β-glucan in comparison to endosperm, which contained 70%. A study investigating the individual cytolytic composition of different barley varieties found that cytolytic malt modifications have a lower β-glucan content. This work confirms that breeding progress reduces β-glucan content in barley [135]. These variances can cause huge inconsistencies in reported therapeutic effects. These two issues can potentially be resolved by producing standardized methods for the extraction of β-glucans categorized by source, as the location of β-glucans is different in each. At present, there is no standardized extraction method.
Furthermore, very few reports state the way in which β-glucans are extracted and the final concentration yield. A potential solution to this is that all β-glucans should be fully characterized in terms of structure, molecular weight and degree of purity. Therefore, activity can be compared, receptor pathways defined and mechanisms of action elucidated.
Finally, disease targets are also very broad. In categorizing and defining structure and disease targets, clinical effects can be better understood. Optimal routes of administration and general applications such as length of treatment and dose also vary in both preclinical and clinical trials, leading to conflicting findings [136]. These too need to be defined for comparison to overcome inconsistences, as there is huge enthusiasm for the clinical use of β-glucans.
5.3. Animal Health
β-glucans administered as adjuvants in vaccines have also shown promise in animal models. In vivo models have shown upregulated macrophage and dendritic cell recruitment and maturation. Antigen-specific CD8+ T-cell responses were also increased. Delivery by various routes of administration showed similar activity [137][138].
There are constant influxes of studies demonstrating the immune modulating properties effects of β-glucans. There is also a ban on the use of antibiotics administered to farmed animals for growth and immune promoting purposes. The use of β-glucans as an alternative is increasing [139]. In fisheries, it is tedious and costly to vaccinate fish to improve immunity and immune strength against pathogens; one of the major benefits of food component immune modulators is that they can be administered as food and still achieve the same effects. Studies have shown that feeding β-glucans to healthy fish has increased non-specific and specific immunity levels and increased protection against bacterial infections [140].
Algae β-glucans have demonstrated improved immunity in dogs [141]. In mice, the bioactives had significant effects as a prophylactic treatment to reduce anthrax infection and inhibition of cancer cells through stimulation of cytokines [142][143]. Sheep oral supplementation to ewes had positive effects on reproductive performance, growth rate and body composition [144]. Lambs, when administered β-glucans, had a significant increase in phagocytic and respiratory burst activities [145][146]. In pigs, there was an improvement in health status [139] and when administered to piglets, there was a reduction in the susceptibility of newborn piglets to enterotoxigenic bacterial infection [147]. When chicks are administered dietary supplementation with yeast derived β-glucans, Salmonella colonization in the cecum was reduced [148]; this effect was also observed in turkeys [149]. With these studies and many more, β-glucans are proving to be an attractive safe alternative to antibiotic administration.
5.4. Applications of β-Glucans in the Food Industry
β-glucans are added to food products for a variety of reasons, most often as texture enhancers [150]. β-glucans originating from barley have been added as a thickener to liquid products as well as a source of dietary fiber [21]. β-glucan have also been demonstrated to increase water absorption [151], be a fat substitute [152] and act as a stabilizer in the formation of foam and emulsion. [153]. When used as a fat substitute in meat, it improved texture and retained moisture [152]. When added to food, β-glucans can enhance texture and replace fats but also have beneficial therapeutic effects against metabolic conditions. β-glucans are not digestible by enzymes in the human gastrointestinal tract, and therefore, when ingested in food, they behave as soluble dietary fibers [154]. Highly viscous β-glucans have the ability to lower cholesterol and improve glycemic control as well as satiety control [117]. There is also evidence to suggest that when eaten, β-glucans interact and positively influence the gut microbiome [155].
The circular economy repurposes residues or by-products into raw materials for other processes, with the aim of reducing waste. Brewers spent grain (BSG) is an abundant by-product from the brewing industry. It contains many bioactives and is a source of 1,3 1,4 β-glucans. Previously, BSG was mainly used in animal nutrition. However, β-glucans can be extracted from BSG and used for human nutrition or included in food with beneficial effects, including reduction in the postprandial glycemic response by up to 50% [156].
6. Conclusions and Perspectives
β-glucans are a diverse class of complex polysaccharides that can be found in an abundance of sources and are classified by source as cereal or non-cereal, based on differences in their branching patterns. These structural differences, caused in part by their source, and the extraction and purification methods used, confer different effect profiles on members of the β-glucans family. There is a significant body of literature attesting to their therapeutic potential, which vary from anti-tumor and immunomodulatory effects described predominantly with mushroom derived β-glucans, to metabolic effects such as improved glycemic control and cholesterol-lowering effects with cereal β-glucans, to probiotic properties.
While over 200 registered clinical trials of β-glucans clearly attest to the interest in these compounds, significant barriers exist to realizing their therapeutic effects. A key issue is the fact that the structure versus function profile remains incompletely understood. Addressing these barriers will require optimization of isolation and purification procedures, careful characterization of the relationship between specific variations in β-glucan structure and their effect profile. This approach would facilitate a greater understanding β-glucan, enabling identification of the most promising compounds for clinical testing, and should realize the promise of this intriguing class of compounds.
References
- Murphy, E.J.; Rezoagli, E.; Major, I.; Rowan, N.J.; Laffey, J.G. β-Glucan Metabolic and Immunomodulatory Properties and Potential for Clinical Application. J. Fungi 2020, 6, 356.
- Wang, Q.; Sheng, X.; Shi, A.; Hu, H.; Yang, Y.; Liu, L.; Fei, L.; Liu, H. β-Glucans: Relationships between modification, conformation and functional activities. Molecules 2017, 22, 257.
- Lordan, S.; Ross, R.P.; Stanton, C. Marine bioactives as functional food ingredients: Potential to reduce the incidence of chronic diseases. Mar. Drugs 2011, 9, 1056–1100.
- Ahnen, R.T.; Jonnalagadda, S.S.; Slavin, J.L. Role of plant protein in nutrition, wellness, and health. Nutr. Rev. 2019, 77, 735–747.
- Kalra, E.K. Nutraceutical-Definition and introduction. AAPS PharmSci 2003, 5, 27.
- Frank, J.; Fukagawa, N.K.; Bilia, A.R.; Johnson, E.J.; Kwon, O.; Prakash, V.; Miyazawa, T.; Clifford, M.N.; Kay, C.D.; Crozier, A.; et al. Terms and nomenclature used for plant-derived components in nutrition and related research: Efforts toward harmonization. Nutr. Rev. 2020, 78, 451–458.
- Santos, D.I.; Saraiva, J.M.A.; Vicente, A.A.; Moldão-Martins, M. Methods for determining bioavailability and bioaccessibility of bioactive compounds and nutrients. In Innovative Thermal and Non-Thermal Processing, Bioaccessibility and Bioavailability of Nutrients and Bioactive Compounds; Elsevier: Amsterdam, The Netherlands, 2019; pp. 23–54.
- Ulmius, M.; Önning, G.; Nilsson, L. Solution behavior of barley β-glucan as studied with asymmetrical flow field-flow fractionation. Food Hydrocoll. 2012, 26, 175–180.
- Rodrigues, M.V.; Zanuzzo, F.S.; Koch, J.F.A.; de Oliveira, C.A.F.; Sima, P.; Vetvicka, V. Development of Fish Immunity and the Role of β-Glucan in Immune Responses. Molecules 2020, 25, 5378.
- Byrne, K.A.; Loving, C.L.; McGill, J.L. Innate Immunomodulation in Food Animals: Evidence for Trained Immunity? Front. Immunol. 2020, 11.
- Ahmad, A.; Anjum, F.M.; Zahoor, T.; Nawaz, H.; Dilshad, S.M.R. Beta glucan: A valuable functional ingredient in foods. Crit. Rev. Food Sci. Nutr. 2012, 52, 201–212.
- Vetvicka, V.; Teplyakova, T.V.; Shintyapina, A.B.; Korolenko, T.A. Effects of medicinal fungi-derived β-glucan on tumor progression. J. Fungi 2021, 7, 250.
- Pillemer, L.; Blum, L.; Lepow, I.H.; Ross, O.A.; Todd, E.W.; Wardlaw, A.C. The properdin system and immunity: I. Demonstration and isolation of a new serum protein, properdin, and its role in immune phenomena. Science 1954, 120, 279–285.
- Chihara, G.; Maeda, Y.; Hamuro, J.; Sasaki, T.; Fukuoka, F. Inhibition of mouse sarcoma 180 by polysaccharides from Lentinus edodes (Berk.) sing. Nature 1969, 222, 687–688.
- Takeshita, K.; Saito, N.; Sato, Y.; Maruyama, M.; Sunagawa, M.; Habu, H.; Endo, M. Diversity of complement activation by lentinan, an antitumor polysaccharide, in gastric cancer patients. Nippon. Geka Gakkai Zasshi 1991, 92, 5–11.
- Kimura, Y.; Tojima, H.; Fukase, S.; Takeda, K. Clinical evaluation of sizofilan as assistant immunotherapy in treatment of head and neck cancer. Acta Otolaryngol. 1994, 114, 192–195.
- Matsuoka, H.; Seo, Y.; Wakasugp, H.; Saito, T.; Tomoda, H. Lentinan potentiates immunity and prolongs the survival time of some patients. Anticancer Res. 1997, 17, 2751–2755. Available online: https://europepmc.org/article/med/9252710 (accessed on 18 May 2021).
- Ina, K.; Kataoka, T.; Ando, T. The Use of Lentinan for Treating Gastric Cancer. Anticancer Agents Med. Chem. 2013, 13, 681–688.
- Efficacy and Tolerability of ABBC1 in Volunteers Receiving the Influenza or COVID-19 Vaccine-Full Text View-ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT04798677?term=bETA-GLUCAN&cond=Covid19&draw=2&rank=1 (accessed on 21 May 2021).
- Henrion, M.; Francey, C.; Lê, K.A.; Lamothe, L. Cereal B-glucans: The impact of processing and how it affects physiological responses. Nutrients 2019, 11, 1729.
- Zhang, H.; Zhang, N.; Xiong, Z.; Wang, G.; Xia, Y.; Lai, P.; Ai, L. Structural characterization and rheological properties of β-D-glucan from hull-less barley (Hordeum vulgare L. var. nudum Hook. f.). Phytochemistry 2018, 155, 155–163.
- Friedman, M. Mushroom polysaccharides: Chemistry and antiobesity, antidiabetes, anticancer, and antibiotic properties in cells, rodents, and humans. Foods 2016, 5, 80.
- Han, B.; Baruah, K.; Cox, E.; Vanrompay, D.; Bossier, P. Structure-Functional Activity Relationship of β-Glucans From the Perspective of Immunomodulation: A Mini-Review. Front. Immunol. 2020, 11, 658.
- Kaur, R.; Sharma, M.; Ji, D.; Xu, M.; Agyei, D. Structural features, modification, and functionalities of beta-glucan. Fibers 2020, 8, 1.
- Rieder, A.; Grimmer, S.; Kolset, S.O.; Michaelsen, T.E.; Knutsen, S.H. Cereal β-glucan preparations of different weight average molecular weights induce variable cytokine secretion in human intestinal epithelial cell lines. Food Chem. 2011, 128, 1037–1043.
- Jin, Y.; Li, P.; Wang, F. β-glucans as potential immunoadjuvants: A review on the adjuvanticity, structure-activity relationship and receptor recognition properties. Vaccine 2018, 36, 5235–5244.
- Du, B.; Meenu, M.; Liu, H.; Xu, B. A concise review on the molecular structure and function relationship of β-glucan. Int. J. Mol. Sci. 2019, 20, 4032.
- Zhan, X.B.; Lin, C.C.; Zhang, H.T. Recent advances in curdlan biosynthesis, biotechnological production, and applications. Appl. Microbiol. Biotechnol. 2012, 93, 525–531.
- Da Silva, B.P.; Silva, G.M.; Mendes, T.P.; Parente, J.P. Structural characteristics of a bioactive polysaccharide from Sorghum arundinaceum. Z. Naturforsch.-Sect. C J. Biosci. 2003, 58, 342–346.
- Ermawar, R.A.; Collins, H.M.; Byrt, C.S.; Betts, N.S.; Henderson, M.; Shirley, N.J.; Schwerdt, J.; Lahnstein, J.; Fincher, G.B.; Burton, R.A. Distribution, structure and biosynthetic gene families of (1,3;1,4)-β-glucan in Sorghum bicolor. J. Integr. Plant Biol. 2015, 57, 429–445.
- Lazaridou, A.; Biliaderis, C.G. Molecular aspects of cereal β-glucan functionality: Physical properties, technological applications and physiological effects. J. Cereal Sci. 2007, 46, 101–118.
- Jin, Y.; Cai, L.; Yang, Q.; Luo, Z.; Liang, L.; Liang, Y.; Wu, B.; Ding, L.; Zhang, D.; Xu, X.; et al. Anti-leukemia activities of selenium nanoparticles embedded in nanotube consisted of triple-helix β-D-glucan. Carbohydr. Polym. 2020, 240, 116329.
- Lyu, F.; Xu, X.; Zhang, L. Natural polysaccharides with different conformations: Extraction, structure and anti-tumor activity. J. Mater. Chem. B 2020, 8, 9652–9667.
- Zheng, X.; Lu, F.; Xu, X.; Zhang, L. Extended chain conformation of β-glucan and its effect on antitumor activity. J. Mater. Chem. B 2017, 5, 5623–5631.
- Stier, H.; Ebbeskotte, V.; Gruenwald, J. Immune-modulatory effects of dietary Yeast Beta-1,3/1,6-D-glucan. Nutr. J. 2014, 13, 38.
- Kim, H.S.; Hong, J.T.; Kim, Y.; Han, S.-B. Stimulatory Effect of β-glucans on Immune Cells. Immune Netw. 2011, 11, 191.
- Zargarzadeh, M.; Amaral, A.J.R.; Custódio, C.A.; Mano, J.F. Biomedical applications of laminarin. Carbohydr. Polym. 2020, 232.
- Desamero, M.J.; Kakuta, S.; Chambers, J.K.; Uchida, K.; Hachimura, S.; Takamoto, M.; Nakayama, J.; Nakayama, H.; Kyuwa, S. Orally administered brown seaweed-derived β-glucan effectively restrained development of gastric dysplasia in A4gnt KO mice that spontaneously develop gastric adenocarcinoma. Int. Immunopharmacol. 2018, 60, 211–220.
- Hong, S.J.; Ahn, M.H.; Sangshetti, J.; Choung, P.H.; Arote, R.B. Sugar-based gene delivery systems: Current knowledge and new perspectives. Carbohydr. Polym. 2018, 181, 1180–1193.
- Sellimi, S.; Maalej, H.; Rekik, D.M.; Benslima, A.; Ksouda, G.; Hamdi, M.; Sahnoun, Z.; Li, S.; Nasri, M.; Hajji, M. Antioxidant, antibacterial and in vivo wound healing properties of laminaran purified from Cystoseira barbata seaweed. Int. J. Biol. Macromol. 2018, 119, 633–644.
- Bobadilla, F.; Rodriguez-Tirado, C.; Imarai, M.; Galotto, M.J.; Andersson, R. Soluble β-1,3/1,6-glucan in seaweed from the southern hemisphere and its immunomodulatory effect. Carbohydr. Polym. 2013, 92, 241–248.
- Krajčovič, J.; Vesteg, M.; Schwartzbach, S.D. Euglenoid flagellates: A multifaceted biotechnology platform. J. Biotechnol. 2015, 202, 135–145.
- Evans, M.; Falcone, P.H.; Crowley, D.C.; Sulley, A.M.; Campbell, M.; Zakaria, N.; Lasrado, J.A.; Fritz, E.P.; Herrlinger, K.A. Effect of a Euglena gracilis fermentate on immune function in healthy, active adults: A randomized, double-blind, placebo-controlled trial. Nutrients 2019, 11, 2926.
- Russo, R.; Barsanti, L.; Evangelista, V.; Frassanito, A.M.; Longo, V.; Pucci, L.; Penno, G.; Gualtieri, P. Euglena gracilis paramylon activates human lymphocytes by upregulating pro-inflammatory factors. Food Sci. Nutr. 2017, 5, 205–214.
- Sikora, P.; Tosh, S.M.; Brummer, Y.; Olsson, O. Identification of high β-glucan oat lines and localization and chemical characterization of their seed kernel β-glucans. Food Chem. 2013, 137, 83–91.
- Daou, C.; Zhang, H. Oat Beta-Glucan: Its Role in Health Promotion and Prevention of Diseases. Compr. Rev. Food Sci. Food Saf. 2012, 11, 355–365.
- Zhu, F.; Du, B.; Xu, B. A critical review on production and industrial applications of beta-glucans. Food Hydrocoll. 2016, 52, 275–288.
- Du, B.; Zhu, F.; Xu, B. β-Glucan extraction from bran of hull-less barley by accelerated solvent extraction combined with response surface methodology. J. Cereal Sci. 2014, 59, 95–100.
- Yan, J.K.; Wang, W.Q.; Wu, J.Y. Recent advances in Cordyceps sinensis polysaccharides: Mycelial fermentation, isolation, structure, and bioactivities: A review. J. Funct. Foods 2014, 6, 33–47.
- Bhanja, S.K.; Rout, D.; Patra, P.; Sen, I.K.; Nandan, C.K.; Islam, S.S. Water-insoluble glucans from the edible fungus Ramaria botrytis. Bioact. Carbohydr. Diet. Fibre 2014, 3, 52–58.
- Kim, Y.W.; Kim, K.H.; Choi, H.J.; Lee, D.S. Anti-diabetic activity of β-glucans and their enzymatically hydrolyzed oligosaccharides from Agaricus blazei. Biotechnol. Lett. 2005, 27, 483–487.
- Brennan, C.S.; Cleary, L.J. The potential use of cereal (1→3,1→4)-β-d-glucans as functional food ingredients. J. Cereal Sci. 2005, 42, 1–13.
- Zhu, F.; Du, B.; Bian, Z.; Xu, B. β-Glucans from edible and medicinal mushrooms: Characteristics, physicochemical and biological activities. J. Food Compos. Anal. 2015, 41, 165–173.
- Kono, H.; Kondo, N.; Hirabayashi, K.; Ogata, M.; Totani, K.; Ikematsu, S.; Osada, M. NMR spectroscopic structural characterization of a water-soluble β-(1 → 3, 1 → 6)-glucan from Aureobasidium pullulans. Carbohydr. Polym. 2017, 174, 876–886.
- Murphy, E.J.; Masterson, C.; Rezoagli, E.; O’Toole, D.; Major, I.; Stack, G.D.; Lynch, M.; Laffey, J.G.; Rowan, N.J. β-Glucan extracts from the same edible shiitake mushroom Lentinus edodes produce differential in-vitro immunomodulatory and pulmonary cytoprotective effects—Implications for coronavirus disease (COVID-19) immunotherapies. Sci. Total Environ. 2020, 732, 139330.
- McCleary, B.V.; Draga, A. Measurement of β-Glucan in mushrooms and mycelial products. J. AOAC Int. 2016, 99, 364–373.
- Liu, H.; Li, Y.; You, M.; Liu, X. Comparison of physicochemical properties of β-glucans extracted from hull-less barley bran by different methods. Int. J. Biol. Macromol. 2021, 182, 1192–1199.
- Kim, H.J.; Kim, H.J. Physicochemical characteristics and in vitro bile acid binding and starch digestion of β-glucans extracted from different varieties of Jeju barley. Food Sci. Biotechnol. 2017, 26, 1501–1510.
- Karimi, R.; Azizi, M.H.; Xu, Q. Effect of different enzymatic extractions on molecular weight distribution, rheological and microstructural properties of barley bran β-glucan. Int. J. Biol. Macromol. 2019, 126, 298–309.
- Benito-Román, Ó.; Alvarez, V.H.; Alonso, E.; Cocero, M.J.; Saldaña, M.D.A. Pressurized aqueous ethanol extraction of β-glucans and phenolic compounds from waxy barley. Food Res. Int. 2015, 75, 252–259.
- Chen, H.; Nie, Q.; Xie, M.; Yao, H.; Zhang, K.; Yin, J.; Nie, S. Protective effects of β-glucan isolated from highland barley on ethanol-induced gastric damage in rats and its benefits to mice gut conditions. Food Res. Int. 2019, 122, 157–166.
- Zielke, C.; Kosik, O.; Ainalem, M.L.; Lovegrove, A.; Stradner, A.; Nilsson, L. Characterization of cereal β-glucan extracts from oat and barley and quantification of proteinaceous matter. PLoS ONE 2017, 12, e0172034.
- Harasym, J.; Zyła, E.; Dziendzikowska, K.; Gromadzka-Ostrowska, J. Proteinaceous residue removal from oat β-glucan extracts obtained by alkalinewater extraction. Molecules 2019, 24, 1729.
- Yoo, H.U.; Ko, M.J.; Chung, M.S. Hydrolysis of beta-glucan in oat flour during subcritical-water extraction. Food Chem. 2020, 308.
- Karp, S.; Wyrwisz, J.; Kurek, M.A. The impact of different levels of oat β-glucan and water on gluten-free cake rheology and physicochemical characterisation. J. Food Sci. Technol. 2020, 57, 3628–3638.
- Yoshida, T.; Honda, Y.; Tsujimoto, T.; Uyama, H.; Azuma, J.I. Selective isolation of β-glucan from corn pericarp hemicelluloses by affinity chromatography on cellulose column. Carbohydr. Polym. 2014, 111, 538–542.
- Li, H.; Wang, Y.; Wang, C.; Zhang, S.; Li, S.; Zhou, G.; Wang, S.; Zhang, J. Extraction, selenylation modification and antitumor activity of the glucan from Castanea mollissima Blume. Glycoconj. J. 2017, 34, 207–217.
- Dong, Z.; Zhang, M.; Li, H.; Zhan, Q.; Lai, F.; Wu, H. Structural characterization and immunomodulatory activity of a novel polysaccharide from Pueraria lobata (Willd.) Ohwi root. Int. J. Biol. Macromol. 2020, 154, 1556–1564.
- Fazio, A.; la Torre, C.; Caroleo, M.C.; Caputo, P.; Plastina, P.; Cione, E. Isolation and purification of glucans from an Italian cultivar of Ziziphus jujuba Mill. And in vitro effect on skin repair. Molecules 2020, 25, 968.
- Tejedor-Calvo, E.; Morales, D.; Marco, P.; Sánchez, S.; Garcia-Barreda, S.; Smiderle, F.R.; Iacomini, M.; Villalva, M.; Santoyo, S.; Soler-Rivas, C. Screening of bioactive compounds in truffles and evaluation of pressurized liquid extractions (PLE) to obtain fractions with biological activities. Food Res. Int. 2020, 132, 109054.
- Alonso-Simón, A.; Encina, A.E.; Seyama, T.; Kondo, T.; García-Angulo, P.; Álvarez, J.M.; Acebes, J.L.; Hayashi, T. Purification and characterization of a soluble β-1,4-glucan from bean (Phaseolus vulgaris L.)-cultured cells dehabituated to dichlobenil. Planta 2013, 237, 1475–1482.
- Fazio, A.; Iacopetta, D.; la Torre, C.; Ceramella, J.; Muià, N.; Catalano, A.; Carocci, A.; Sinicropi, M.S. Finding solutions for agricultural wastes: Antioxidant and antitumor properties of pomegranate Akko peel extracts and β-glucan recovery†. Food Funct. 2018, 9, 6619–6632.
- Muñoz-Almagro, N.; Gilbert-López, B.; Pozuelo-Rollón, M.C.; García-Fernandez, Y.; Almeida, C.; Villamiel, M.; Mendiola, J.A.; Ibáñez, E. Exploring the microalga euglena cantabrica by pressurized liquid extraction to obtain bioactive compounds. Mar. Drugs 2020, 18, 308.
- Rajauria, G.; Ravindran, R.; Garcia-Vaquero, M.; Rai, D.K.; Sweeney, T.; O’Doherty, J. Molecular characteristics and antioxidant activity of laminarin extracted from the seaweed species Laminaria hyperborea, using hydrothermal-assisted extraction and a multi-step purification procedure. Food Hydrocoll. 2021, 112, 106332.
- Borchani, C.; Fonteyn, F.; Jamin, G.; Paquot, M.; Blecker, C.; Thonart, P. Enzymatic process for the fractionation of baker’s yeast cell wall (Saccharomyces cerevisiae). Food Chem. 2014, 163, 108–113.
- Kupetz, M.; Procopio, S.; Sacher, B.; Becker, T. Critical review of the methods of β-glucan analysis and its significance in the beer filtration process. Eur. Food Res. Technol. 2015, 241, 725–736.
- Dhewantara, F.X.R. Cholesterol-lowering effect of beta glucan extracted from saccharomyces cerevisiae in rats. Sci. Pharm. 2016, 84, 153–165.
- Amer, E.M.; Saber, S.H.; Markeb, A.A.; Elkhawaga, A.A.; Mekhemer, I.M.A.; Zohri, A.N.A.; Abujamel, T.S.; Harakeh, S.; Abd-Allah, E.A. Enhancement of β-glucan biological activity using a modified acid-base extraction method from saccharomyces cerevisiae. Molecules 2021, 26, 2113.
- Bzducha-Wróbel, A.; Blłazejak, S.; Kawarska, A.; Stasiak-Rózańska, L.; Gientka, I.; Majewska, E. Evaluation of the efficiency of different disruption methods on yeast cell wall preparation for β-glucan isolation. Molecules 2014, 19, 20941–20961.
- Upadhyay, T.K.; Fatima, N.; Sharma, D.; Saravanakumar, V.; Sharma, R. Preparation and characterization of beta-glucan particles containing a payload of nanoembedded rifabutin for enhanced targeted delivery to macrophages. EXCLI J. 2017, 16, 210–228.
- Abreu, H.; Zavadinack, M.; Smiderle, F.R.; Cipriani, T.R.; Cordeiro, L.M.C.; Iacomini, M. Polysaccharides from Pleurotus eryngii: Selective extraction methodologies and their modulatory effects on THP-1 macrophages. Carbohydr. Polym. 2021, 252.
- Rodríguez-Seoane, P.; González-Muñoz, M.J.; Falqué, E.; Domínguez, H. Pressurized hot water extraction of β-glucans from Cantharellus tubaeformis. Electrophoresis 2018, 39, 1892–1898.
- Carbonero, E.R.; Ruthes, A.C.; Freitas, C.S.; Utrilla, P.; Gálvez, J.; da Silva, E.V.; Sassaki, G.L.; Gorin, P.A.J.; Iacomini, M. Chemical and biological properties of a highly branched β-glucan from edible mushroom Pleurotus sajor-caju. Carbohydr. Polym. 2012, 90, 814–819.
- Palanisamy, M.; Aldars-García, L.; Gil-Ramírez, A.; Ruiz-Rodríguez, A.; Marín, F.R.; Reglero, G.; Soler-Rivas, C. Pressurized water extraction of β-glucan enriched fractions with bile acids-binding capacities obtained from edible mushrooms. Biotechnol. Prog. 2014, 30, 391–400.
- Umaña, M.; Eim, V.; Garau, C.; Rosselló, C.; Simal, S. Ultrasound-assisted extraction of ergosterol and antioxidant components from mushroom by-products and the attainment of a β-glucan rich residue. Food Chem. 2020, 332.
- Alzorqi, I.; Sudheer, S.; Lu, T.J.; Manickam, S. Ultrasonically extracted β-D-glucan from artificially cultivated mushroom, characteristic properties and antioxidant activity. Ultrason. Sonochem. 2017, 35, 531–540.
- Abreu, H.; Simas, F.F.; Smiderle, F.R.; Sovrani, V.; Dallazen, J.L.; Maria-Ferreira, D.; Werner, M.F.; Cordeiro, L.M.C.; Iacomini, M. Gelling functional property, anti-inflammatory and antinociceptive bioactivities of β-D-glucan from the edible mushroom Pholiota nameko. Int. J. Biol. Macromol. 2019, 122, 1128–1135.
- Szwengiel, A.; Stachowiak, B. Deproteinization of water-soluble ß-glucan during acid extraction from fruiting bodies of Pleurotus ostreatus mushrooms. Carbohydr. Polym. 2016, 146, 310–319.
- Li, J.; Cai, C.; Zheng, M.; Hao, J.; Wang, Y.; Hu, M.; Fan, L.; Yu, G. Alkaline extraction, structural characterization, and bioactivities of (1→6)-β-d-glucan from lentinus edodes. Molecules 2019, 24, 1610.
- Rieder, A.; Knutsen, S.H.; Fernandez, A.S.; Ballance, S. At a high dose even partially degraded beta-glucan with decreased solubility significantly reduced the glycaemic response to bread. Food Funct. 2019, 10, 1529–1539.
- Panneerselvam, D.; Budh, D.P. Peyer Patches. In Encyclopedia of Cancer; Springer: Berlin/Heidelberg, Germany, 2011; p. 2831.
- Batbayar, S.; Lee, D.H.; Kim, H.W. Immunomodulation of fungal β-glucan in host defense signaling by dectin-1. Biomol. Ther. 2012, 20, 433–445.
- Reboldi, A.; Cyster, J.G. Peyer’s patches: Organizing B-cell responses at the intestinal frontier. Immunol. Rev. 2016, 271, 230–245.
- Mahla, R.S.; Reddy, M.C.; Prasad, D.V.R.; Kumar, H. Sweeten PAMPs: Role of sugar complexed PAMPs in innate immunity and vaccine biology. Front. Immunol. 2013, 4.
- Camilli, G.; Tabouret, G.; Quintin, J. The Complexity of Fungal β-Glucan in Health and Disease: Effects on the Mononuclear Phagocyte System. Front. Immunol. 2018, 9.
- Kolarich, D.; Lepenies, B.; Seeberger, P.H. Glycomics, glycoproteomics and the immune system. Curr. Opin. Chem. Biol. 2012, 16, 214–220.
- Rabinovich, G.A.; van Kooyk, Y.; Cobb, B.A. Glycobiology of immune responses. Ann. N. Y. Acad. Sci. 2012, 1253, 1–15.
- Patin, E.C.; Thompson, A.; Orr, S.J. Pattern recognition receptors in fungal immunity. Semin. Cell Dev. Biol. 2019, 89, 24–33.
- Tang, J.; Lin, G.; Langdon, W.Y.; Tao, L.; Zhang, J. Regulation of C-type lectin receptor-mediated antifungal immunity. Front. Immunol. 2018, 9, 123.
- Cohen-Kedar, S.; Baram, L.; Elad, H.; Brazowski, E.; Guzner-Gur, H.; Dotan, I. Human intestinal epithelial cells respond to β-glucans via Dectin-1 and Syk. Eur. J. Immunol. 2014, 44, 3729–3740.
- Branzk, N.; Lubojemska, A.; Hardison, S.E.; Wang, Q.; Gutierrez, M.G.; Brown, G.D.; Papayannopoulos, V. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat. Immunol. 2014, 15, 1017–1025.
- Li, P.; Wang, F. Polysaccharides: Candidates of promising vaccine adjuvants. Drug Discov. Ther. 2015, 9, 88–93.
- Kardani, K.; Bolhassani, A.; Shahbazi, S. Prime-boost vaccine strategy against viral infections: Mechanisms and benefits. Vaccine 2016, 34, 413–423.
- Haas, T.; Heidegger, S.; Wintges, A.; Bscheider, M.; Bek, S.; Fischer, J.C.; Eisenkolb, G.; Schmickl, M.; Spoerl, S.; Peschel, C.; et al. Card9 controls Dectin-1-induced T-cell cytotoxicity and tumor growth in mice. Eur. J. Immunol. 2017, 47, 872–879.
- Vorup-Jensen, T.; Jensen, R.K. Structural immunology of complement receptors 3 and 4. Front. Immunol. 2018, 9, 2716.
- Bajic, G.; Yatime, L.; Sim, R.B.; Vorup-Jensen, T.; Andersen, G.R. Structural insight on the recognition of surface-bound opsonins by the integrin i domain of complement receptor 3. Proc. Natl. Acad. Sci. USA 2013, 110, 16426–16431.
- Geller, A.; Shrestha, R.; Yan, J. Yeast-derived β-glucan in cancer: Novel uses of a traditional therapeutic. Int. J. Mol. Sci. 2019, 20, 3618.
- Vera, J.; Fenutria, R.; Cañadas, O.; Figueras, M.; Mota, R.; Sarrias, M.R.; Williams, D.L.; Casals, C.; Yelamos, J.; Lozano, F. The CD5 ectodomain interacts with conserved fungal cell wall components and protects from zymosan-induced septic shock-like syndrome. Proc. Natl. Acad. Sci. USA 2009, 106, 1506–1511.
- Yang, C.; Gao, J.; Dong, H.; Zhu, P.F.; Wang, Z.G.; Jiang, J.X. Expressions of scavenger receptor, CD14 and protective mechanisms of carboxymethyl-β2-1, 3-glucan in posttraumatic endotoxemia in mice. J. Trauma-Inj. Infect. Crit. Care 2008, 65, 1471–1477.
- Guo, Y.; Fukuda, T.; Donai, K.; Kuroda, K.; Masuda, M.; Nakamura, S.; Yoneyama, H.; Isogai, E. Leptospiral lipopolysaccharide stimulates the expression of toll-like receptor 2 and cytokines in pig fibroblasts. Anim. Sci. J. 2015, 86, 238–244.
- Medzhitov, R. Recognition of microorganisms and activation of the immune response. Nature 2007, 449, 819–826.
- Adams, E.L.; Rice, P.J.; Graves, B.; Ensley, H.E.; Yu, H.; Brown, G.D.; Gordon, S.; Monteiro, M.A.; Papp-Szabo, E.; Lowman, D.W.; et al. Differential high-affinity interaction of Dectin-1 with natural or synthetic glucans is dependent upon primary structure and is influenced by polymer chain length and side-chain branching. J. Pharmacol. Exp. Ther. 2008, 325, 115–123.
- Maheshwari, G.; Sowrirajan, S.; Joseph, B. Extraction and Isolation of β-Glucan from Grain Sources—A Review. J. Food Sci. 2017, 82, 1535–1545.
- Suzuki, T.; Kusano, K.; Kondo, N.; Nishikawa, K.; Kuge, T.; Ohno, N. Biological Activity of High-Purity β-1,3-1,6-Glucan Derived from the Black Yeast Aureobasidium pullulans: A Literature Review. Nutrients 2021, 13, 242.
- Drozdowski, L.A.; Reimer, R.A.; Temelli, F.; Bell, R.C.; Vasanthan, T.; Thomson, A.B.R. β-Glucan extracts inhibit the in vitro intestinal uptake of long-chain fatty acids and cholesterol and down-regulate genes involved in lipogenesis and lipid transport in rats. J. Nutr. Biochem. 2010, 21, 695–701.
- Tiwari, U.; Cummins, E. Meta-analysis of the effect of β-glucan intake on blood cholesterol and glucose levels. Nutrition 2011, 27, 1008–1016.
- McRorie, J.W.; McKeown, N.M. Understanding the Physics of Functional Fibers in the Gastrointestinal Tract: An Evidence-Based Approach to Resolving Enduring Misconceptions about Insoluble and Soluble Fiber. J. Acad. Nutr. Diet. 2017, 117, 251–264.
- Sima, P.; Vannucci, L.; Vetvicka, V. β-glucans and cholesterol (Review). Int. J. Mol. Med. 2018, 41, 1799–1808.
- Wang, H.; Chen, G.; Li, X.; Zheng, F.; Zeng, X. Yeast β-glucan, a potential prebiotic, showed a similar probiotic activity to inulin. Food Funct. 2020, 11, 10386–10396.
- Ooi, V.E.C.; Liu, F. Immunomodulation and Anti-Cancer Activity of Polysaccharide-Protein Complexes. Curr. Med. Chem. 2012, 7, 715–729.
- Kofuji, K.; Aoki, A.; Tsubaki, K.; Konishi, M.; Isobe, T.; Murata, Y. Antioxidant Activity of β-Glucan. ISRN Pharm. 2012, 2012, 125864.
- Yamada, J.; Hamuro, J.; Hatanaka, H.; Hamabata, K.; Kinoshita, S. Alleviation of seasonal allergic symptoms with superfine β-1,3-glucan: A randomized study. J. Allergy Clin. Immunol. 2007, 119, 1119–1126.
- Quintin, J. Fungal mediated innate immune memory, what have we learned? Semin. Cell Dev. Biol. 2019, 89, 71–77.
- Kamta, J.; Chaar, M.; Ande, A.; Altomare, D.A.; Ait-Oudhia, S. Advancing cancer therapy with present and emerging immuno-oncology approaches. Front. Oncol. 2017, 7, 64.
- Ayeka, P.A. Potential of Mushroom Compounds as Immunomodulators in Cancer Immunotherapy: A Review. Evid.-Based Complementary Altern. Med. 2018, 2018, 7271509.
- Zou, S.; Duan, B.; Xu, X. Inhibition of tumor growth by β-glucans through promoting CD4 + T cell immunomodulation and neutrophil-killing in mice. Carbohydr. Polym. 2019, 213, 370–381.
- Vetvicka, V.; Vannucci, L.; Sima, P.; Richter, J. Beta glucan: Supplement or drug? From laboratory to clinical trials. Molecules 2019, 24, 1251.
- Vetvicka, V.; Vannucci, L.; Sima, P. Β-Glucan As a New Tool in Vaccine Development. Scand. J. Immunol. 2020, 91.
- Temizoz, B.; Kuroda, E.; Ishii, K.J. Vaccine adjuvants as potential cancer immunotherapeutics. Int. Immunol. 2016, 28, 329–338.
- Levitz, S.M.; Huang, H.; Ostroff, G.R.; Specht, C.A. Exploiting fungal cell wall components in vaccines. Semin. Immunopathol. 2015, 37, 199–207.
- Fesel, P.H.; Zuccaro, A. β-glucan: Crucial component of the fungal cell wall and elusive MAMP in plants. Fungal Genet. Biol. 2016, 90, 53–60.
- Bazan, S.B.; Breinig, T.; Schmitt, M.J.; Breinig, F. Heat treatment improves antigen-specific T cell activation after protein delivery by several but not all yeast genera. Vaccine 2014, 32, 2591–2598.
- Miyamoto, N.; Mochizuki, S.; Sakurai, K. Designing an immunocyte-targeting delivery system by use of beta-glucan. Vaccine 2018, 36, 186–189.
- Yuan, H.; Lan, P.; He, Y.; Li, C.; Ma, X. Effect of the modifications on the physicochemical and biological properties of β-glucan-a critical review. Molecules 2020, 25, 57.
- Gastl, M.; Kupetz, M.; Becker, T. Determination of Cytolytic Malt Modification–Part I: Influence of Variety Characteristics. J. Am. Soc. Brew. Chem. 2020, 79, 53–65.
- Castro, E.d.; Calder, P.C.; Roche, H.M. β-1,3/1,6-Glucans and Immunity: State of the Art and Future Directions. Mol. Nutr. Food Res. 2021, 65, 1901071.
- Miyamoto, N.; Mochizuki, S.; Sakurai, K. Enhanced immunostimulation with crosslinked CpG-DNA/β-1,3-glucan nanoparticle through hybridization. Chem. Lett. 2014, 43, 991–993.
- Kobiyama, K.; Temizoz, B.; Kanuma, T.; Ozasa, K.; Momota, M.; Yamamoto, T.; Aoshi, T.; Kuroda, E.; Ishii, K.J. Species-dependent role of type I IFNs and IL-12 in the CTL response induced by humanized CpG complexed with β-glucan. Eur. J. Immunol. 2016, 46, 1142–1151.
- Vetvicka, V.; Vannucci, L.; Sima, P. The Effects of β-Glucan on Pig Growth and Immunity. Open Biochem. J. 2014, 1, 89–93.
- Pogue, R.; Murphy, E.J.; Fehrenbach, G.W.; Rezoagli, E.; Rowan, N.J. Exploiting immunomodulatory properties of β-glucans derived from natural products for improving health and sustainability in aquaculture-farmed organisms: Concise review of existing knowledge, innovation and future opportunities. Curr. Opin. Environ. Sci. Health 2021, 21, 100248.
- Paris, S.; Chapat, L.; Pasin, M.; Lambiel, M.; Sharrock, T.E.; Shukla, R.; Sigoillot-Claude, C.; Bonnet, J.M.; Poulet, H.; Freyburger, L.; et al. β-Glucan-Induced Trained Immunity in Dogs. Front. Immunol. 2020, 11.
- Roudi, R.; Mohammadi, S.R.; Roudbary, M.; Mohsenzadegan, M. Lung cancer and β-glucans: Review of potential therapeutic applications. Investig. New Drugs 2017, 35, 509–517.
- Vetvicka, V.; Vetvickova, J. Comparison of Immunological Effects of Commercially Available β-Glucans. 2014. Available online: http://www.hoajonline.com/journals/pdf/2054-9903-1-1.pdf (accessed on 18 March 2021).
- Zaleska, B.; Milewski, S.; Zabek, K. Impact of Saccharomyces cerevisiae supplementation on reproductive performance, milk yield in ewes and offspring growth. Arch. Anim. Breed. 2015, 58, 79–83.
- Zabek, K.; Milewski, S.; Wójcik, R.; Siwicki, A.K. Effect of β-1,3/1,6-D-glucan in diet on productivity and humoral and cellular defense mechanisms in sheep. Acta Vet. Brno 2013, 82, 141–146.
- Khalkhane, A.S.; Habibian, R. Effect of dietary B-glucan supplementation on humoral and cellular immunologic factors in lambs. Glob. Vet. 2013, 11, 38–43.
- Stuyven, E.; Cox, E.; Vancaeneghem, S.; Arnouts, S.; Deprez, P.; Goddeeris, B.M. Effect of β-glucans on an ETEC infection in piglets. Vet. Immunol. Immunopathol. 2009, 128, 60–66.
- Shao, Y.; Guo, Y.; Wang, Z. β-1,3/1,6-Glucan alleviated intestinal mucosal barrier impairment of broiler chickens challenged with Salmonella enterica serovar Typhimurium. Poult. Sci. 2013, 92, 1764–1773.
- Huff, G.R.; Huff, W.E.; Farnell, M.B.; Rath, N.C.; de los Santos, F.S.; Donoghue, A.M. Bacterial clearance, heterophil function, and hematological parameters of transport-stressed turkey poults supplemented with dietary yeast extract. Poult. Sci. 2010, 89, 447–456.
- Mejía, S.M.V.; de Francisco, A.; Bohrer, B.M. A comprehensive review on cereal β-glucan: Extraction, characterization, causes of degradation, and food application. Crit. Rev. Food Sci. Nutr. 2020, 60, 3693–3704.
- Izydorczyk, M.S.; Dexter, J.E. Barley β-glucans and arabinoxylans: Molecular structure, physicochemical properties, and uses in food products-a Review. Food Res. Int. 2008, 41, 850–868.
- Piñero, M.P.; Parra, K.; Huerta-Leidenz, N.; de Moreno, L.A.; Ferrer, M.; Araujo, S.; Barboza, Y. Effect of oat’s soluble fibre (β-glucan) as a fat replacer on physical, chemical, microbiological and sensory properties of low-fat beef patties. Meat Sci. 2008, 80, 675–680.
- Liu, R.; Wang, N.; Li, Q.; Zhang, M. Comparative studies on physicochemical properties of raw and hydrolyzed oat β-glucan and their application in low-fat meatballs. Food Hydrocoll. 2015, 51, 424–431.
- El Khoury, D.; Cuda, C.; Luhovyy, B.L.; Anderson, G.H. Beta glucan: Health benefits in obesity and metabolic syndrome. J. Nutr. Metab. 2012, 2012, 851362.
- Ryan, P.M.; London, L.E.E.; Bjorndahl, T.C.; Mandal, R.; Murphy, K.; Fitzgerald, G.F.; Shanahan, F.; Ross, R.P.; Wishart, D.S.; Caplice, N.M.; et al. Microbiome and metabolome modifying effects of several cardiovascular disease interventions in apo-E-/- mice. Microbiome 2017, 5, 30.
- Steiner, J.; Procopio, S.; Becker, T. Brewer’s spent grain: Source of value-added polysaccharides for the food industry in reference to the health claims. Eur. Food Res. Technol. 2015, 241, 303–315.
This entry is offline, you can click here to edit this entry!
