The fight against cancer is one of the main challenges for medical research. Recently, nanotechnology has made significant progress, providing possibilities for developing innovative nanomaterials to overcome the common limitations of current therapies. In this context, silver nanoparticles (AgNPs) represent a promising nano-tool able to offer interesting applications for cancer research. Following this path, we combined the silver proprieties with Artemisia arborescens characteristics, producing novel nanoparticles called Artemisia–AgNPs. A “green” synthesis method was performed to produce Artemisia–AgNPs, using Artemisia arborescens extracts. Several biological assays were performed to evaluate the Artemisia–AgNPs anti-cancer properties.
- ancer research
- nanotechnology
- silver nanoparticles
- Artemisia arborescens
- RNA-seq
Note: The following contents are extract from your paper. The entry will be online only after author check and submit it.
1. Introduction
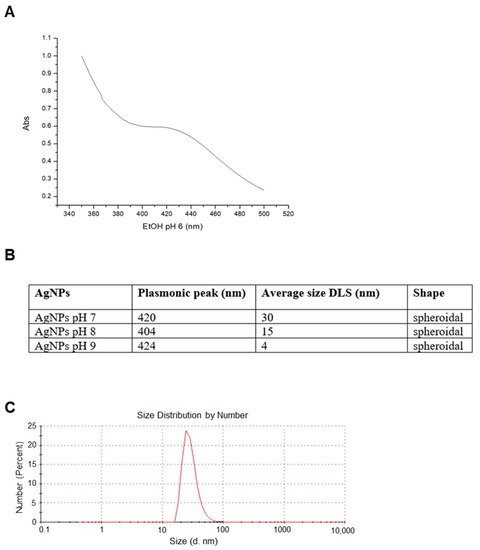
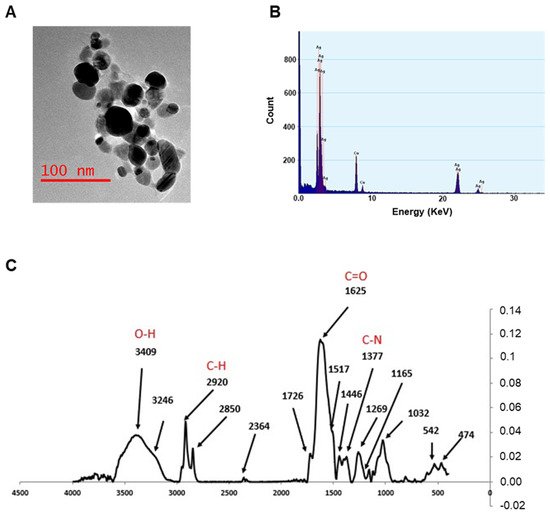
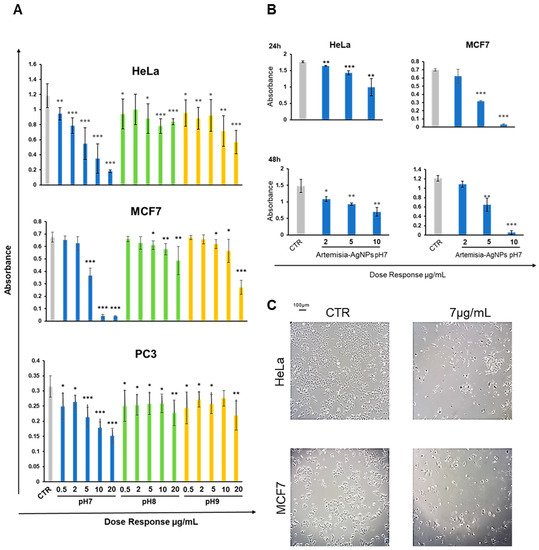
2. Nanoparticles Characterization and XTT Analysis
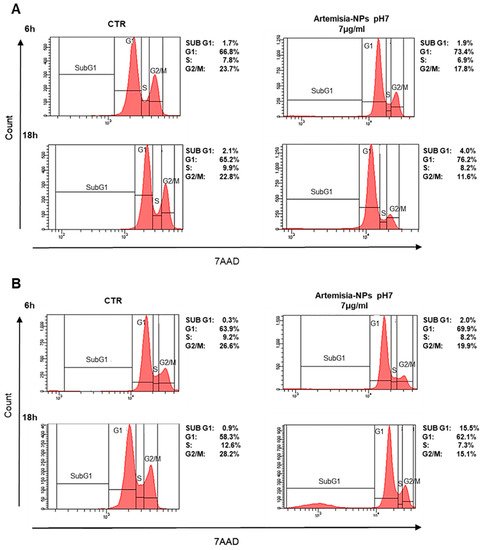
3. Cell Cycle Impact
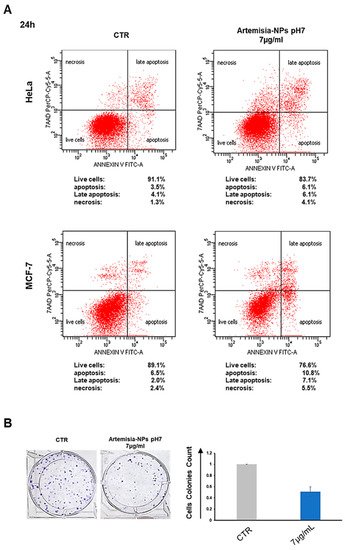
4. Artemisia–AgNPs Induce Apoptosis and Inhibit Colony Formation in Cancer Cells
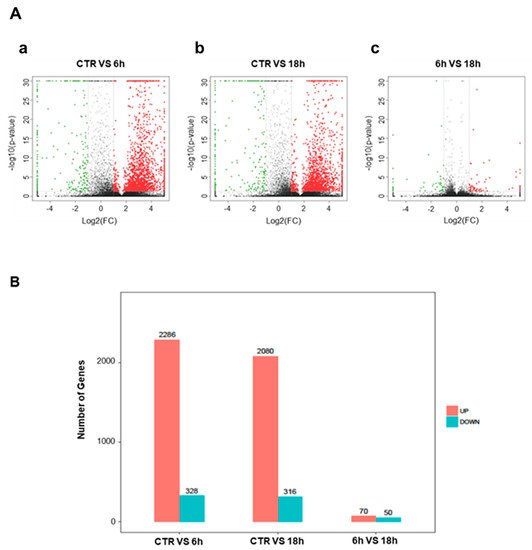
5. Artemisia–AgNPs Impact on Gene Expression
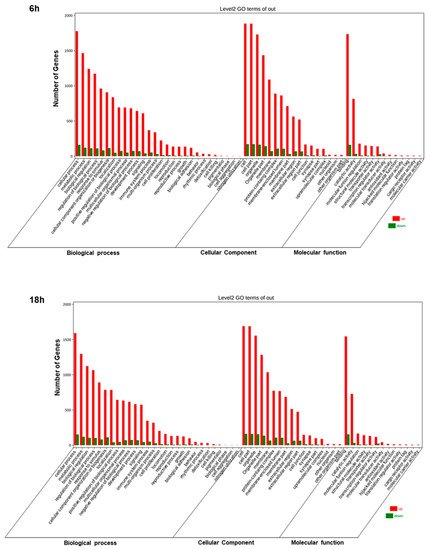
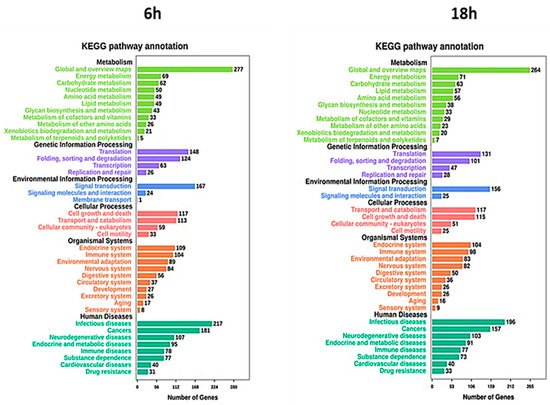
| Category | Term | Description | Gene Count | p Value |
|---|---|---|---|---|
| Biological Process (GO) | GO:0010629 | Negative regulation of gene expression | 12 | 2.33 × 10−12 |
| Biological Process (GO) | GO:0090304 | Nucleic acid metabolic process | 12 | 5.05 × 10−8 |
| Biological Process (GO) | GO:00427969 | DNA demage response, detection of DNA demage | 4 | 9.62 × 10−8 |
| Biological Process (GO) | GO:0042276 | Error-prone translesion synthesis | 4 | 2.46 × 10−6 |
| Biological Process (GO) | GO:0006297 | Nucleotide-excision repair, DNA gap filling | 3 | 3.52 × 10−6 |
| Biological Process (GO) | GO:0070911 | Global genome nucleotide-excision repair | 3 | 4.76 × 10−6 |
| Biological Process (GO) | GO:0000122 | Negative regulation of transcription by RNA polymerase II | 5 | 1.05 × 10−3 |
| KEGG_PATHWAY | Hsa03010 | Ribosome | 11 | 2.96 × 10−20 |
This entry is adapted from the peer-reviewed paper 10.3390/ijms22168621
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424.
- Dobbelstein, M.; Moll, U. Targeting tumour-supportive cellular machineries in anticancer drug development. Nat. Rev. Drug Discov. 2014, 13, 179–196.
- Arruebo, M.; Vilaboa, N.; Sáez-Gutierrez, B.; Lambea, J.; Tres, A.; Valladares, M.; González-Fernández, Á. Assessment of the evolution of cancer treatment therapies. Cancers 2011, 3, 3279–3330.
- Tran, S.; DeGiovanni, P.; Piel, B.; Rai, P. Cancer nanomedicine: A review of recent success in drug delivery. Clin. Transl. Med. 2017, 6, 44.
- Shi, J.; Kantoff, P.W.; Wooster, R.; Farokhzad, O.C. Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 2017, 17, 20–37.
- Ji, S.R.; Liu, C.; Zhang, B.; Yang, F.; Xu, J.; Long, J.; Jin, C.; Fu, D.L.; Ni, Q.X.; Yu, X.J. Carbon nanotubes in cancer diagnosis and therapy. Biochim. Biophys. Acta 2010, 1806, 29–35.
- Gobbo, O.L.; Sjaastad, K.; Radomski, M.W.; Volkov, Y.; Prina-Mello, A. Magnetic Nanoparticles in Cancer Theranostics. Theranostics 2015, 5, 1249–1263.
- Deshpande, P.P.; Biswas, S.; Torchilin, V.P. Current trends in the use of liposomes for tumor targeting. Nanomedicine 2013, 8, 1509–1528.
- Sztandera, K.; Gorzkiewicz, M.; Klajnert-Maculewicz, B. Gold Nanoparticles in Cancer Treatment. Mol. Pharm. 2019, 16, 1–23.
- Grodzinski, P.; Silver, M.; Molnar, L.K. Nanotechnology for cancer diagnostics: Promises and challenges. Expert Rev. Mol. Diagn. 2006, 6, 307–318.
- Medici, S.; Peana, M.; Crisponi, G.; Nurchi, V.M.; Lachowicz, J.I.; Remelli, M.; Zoroddu, M.A. Silver coordination compounds: A new horizon in medicine. Coord. Chem. Rev. 2016, 327, 349–359.
- Foroozandeh, P.; Aziz, A.A. Insight into Cellular Uptake and Intracellular Trafficking of Nanoparticles. Nanoscale Res. Lett. 2018, 13, 339.
- Dayem, A.A.; Hossain, M.K.; Lee, S.B.; Kim, K.; Saha, S.K.; Yang, G.M.; Choi, H.Y.; Cho, S.G. The Role of Reactive Oxygen Species (ROS) in the Biological Activities of Metallic Nanoparticles. Int. J. Mol. Sci. 2017, 18, 120.
- Ahmed, S.; Ahmad, M.; Swami, B.L.; Ikram, S. A review on plants extract mediated synthesis of silver nanoparticles for antimicrobial applications: A green expertise. J. Adv. Res. 2016, 7, 17–28.
- Albanese, A.; Tang, P.S.; Chan, W.C. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng. 2012, 14, 1–16.
- Rajabnia, T.; Meshkini, A. Fabrication of adenosine 5′-triphosphate-capped silver nanoparticles: Enhanced cytotoxicity efficacy and targeting effect against tumor cells. Process Biochem. 2018, 65, 186–196.
- Bhunia, A.K.; Samanta, P.K.; Aich, D.; Saha, S.; Kamilya, T. Biocompatibility study of protein capped and uncapped silver nanoparticles on human hemoglobin. J. Phys. 2015, 48, 235305.
- Raza, M.A.; Kanwal, Z.; Rauf, A.; ASabri, N.; Riaz, S.; Naseem, S. Size- and Shape-Dependent Antibacterial Studies of Silver Nanoparticles Synthesized by Wet Chemical Routes. Nanomaterials 2016, 6, 74.
- Kim, D.H.; Park, J.C.; Jeon, G.E.; Kim, C.S.; Seo, J.H. Effect of the Size and Shape of Silver Nanoparticles on Bacterial Growth and Metabolism by Monitoring Optical Density and Fluorescence Intensity. Biotechnol. Bioprocess Eng. 2017, 22, 210–217.
- Kamali, M.; Ghorashi SA, A.; Asadollahi, M. Controllable Synthesis of Silver Nanoparticles Using Citrate as Complexing Agent: Characterization of Nanopartciles and Effect of pH on Size and Crystallinity. Iran. J. Chem. Chem. Eng. Int. Engl. Ed. 2012, 31, 21–28.
- Avitabile, E.; Senes, N.; D’Avino, C.; Tsamesidis, I.; Pinna, A.; Medici, S.; Pantaleo, A. The potential antimalarial efficacy of hemocompatible silver nanoparticles from Artemisia species against P. falciparum parasite. PLoS ONE 2020, 15, e0238532.
- Martínez-Castañón, G.A.; Niño-Martínez, N.; Martínez-Gutierrez, F.; Martínez-Mendoza, J.R.; Ruiz, F. Synthesis and antibacterial activity of silver nanoparticles with different sizes. J. Nanoparticle Res. 2008, 10, 1343–1348.
- Qin, Y.; Ji, X.; Jing, J.; Liu, H.; Wu, H.; Yang, W. Size control over spherical silver nanoparticles by ascorbic acid reduction. Colloids Surf. A Phys. Eng Asp. 2010, 372, 172–176.
- Urbańska, K.; Pająk, B.; Orzechowski, A.; Sokołowska, J.; Grodzik, M.; Sawosz, E.; Szmidt, M.; Sysa, P. The effect of silver nanoparticles (AgNPs) on proliferation and apoptosis of in ovo cultured glioblastoma multiforme (GBM) cells. Nanoscale Res. Lett. 2015, 10, 98.
- Loutfy, S.A.; Al-Ansary, N.A.; Abdel-Ghani, N.T.; Hamed, A.R.; Mohamed, M.B.; Craik, J.D.; Eldin, T.A.; Abdellah, A.M.; Hussein, Y.; Hasanin, M.T.; et al. Anti-proliferative Activities of Metallic Nanoparticles in an in Vitro Breast Cancer Model. Asian Pac. J. Cancer Prev. 2015, 16, 6039–6046.
- Sur, I.; Altunbek, M.; Kahraman, M.; Culha, M. Interaction of multi-functional silver nanoparticles with living cells. Nanotechnology 2010, 21, 175104.
- Lu, F.; Wu, S.H.; Hung, Y.; Mou, C.Y. Size effect on cell uptake in well-suspended, uniform mesoporous silica nanoparticle. Small 2009, 5, 1408–1413.
- Khorrami, S.; Zarrabi, A.; Khaleghi, M.; Danaei, M.; Mozafari, M.R. Selective cytotoxicity of green synthesized silver nanoparticles against the MCF-7 tumor cell line and their enhanced antioxidant and antimicrobial properties. Int. J. Nanomed. 2018, 13, 8013–8024.
- Al-Sheddi, E.S.; Farshori, N.N.; Al-Oqail, M.M.; Al-Massarani, S.M.; Saquib, Q.; Wahab, R.; Musarrat, J.; Al-Khedhairy, A.A.; Siddiqui, M.A. Anticancer Potential of Green Synthesized Silver Nanoparticles Using Extract of Nepeta deflersiana against Human Cervical Cancer Cells (HeLA). Bioinorg. Chem. Appl. 2018, 2018, 9390784.
- Lee, Y.S.; Kim, D.W.; Lee, Y.H.; Oh, J.H.; Yoon, S.; Choi, M.S.; Lee, S.K.; Kim, J.W.; Lee, K.; Song, C.W. Silver nanoparticles induce apoptosis and G2/M arrest via PKCζ-dependent signaling in A549 lung cells. Arch. Toxicol. 2011, 85, 1529.
- Kovács, D.; Igaz, N.; Keskeny, C.; Bélteky, P.; Tóth, T.; Gáspár, R.; Madarász, D.; Rázga, Z.; Kónya, Z.; Boros, I.M.; et al. Silver nanoparticles defeat p53-positive and p53-negative osteosarcoma cells by triggering mitochondrial stress and apoptosis. Sci. Rep. 2016, 6, 27902.
- Satapathy, S.R.; Mohapatra, P.; Preet, R.; Das, D.; Sarkar, B.; Choudhuri, T.; Wyatt, M.D.; Kundu, C.N. Silver-based nanoparticles induce apoptosis in human colon cancer cells mediated through p53. Nanomedicine 2013, 8, 1307.
- Çiftçi, H.; Türk, M.; Tamer, U.; Karahan, S.; Menemen, Y. Silver nanoparticles: Cytotoxic, apoptotic, and necrotic effects on MCF-7 cells. Turk. J. Biol. 2013, 37, 573–581.
- Deng, L.; Meng, T.; Chen, L.; Wei, W.; Wang, P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct. Target. Ther. 2020, 5, 11.
