Rhodococci are relatively new objects of environmental and industrial biotechnologies. Their metabolic potential for biodegradation and inactivation of complex pollutants, in addition to their mechanisms of stress resistance, are far from being exhausted. However, one should be conscious that some members of this genus are pathogens, and their number is gradually expanding, which clearly limits the practical application of rhodococci.
- actinobacteria
- pathogenicity factors
- adhesion
- autoaggregation
- colonization
- defense against phagocytosis
- adaptive strategies
1. Introduction
Among the stress-tolerant species, actinobacteria of the genus Rhodococcus (phylum Actinobacteria, class Actinomycetia, order Corynebacteriales, family Nocardiaceae) (available online at: https://lpsn.dsmz.de/class/actinomycetia (accessed on 20 July 2021)) stand out due to their greatest variety of degraded xenobiotics and their complete mineralization of chemical pollutants to simple substances using alternative carbon sources (cometabolism), often followed by useful product formation [1][2][3].
A wide range of actinomycetologists focus on the study of polyextremotolerant rhodococci predominant in anthropogenically disturbed biotopes primarily due to their feasible applications in ecobiotechnologies. Although the study of Rhodococcus was, until recently, largely academic, it has now become more applied. This is obviously because, due to their metabolic flexibility, rhodococci do have not many “competitors” in terms of decomposing organic xenobiotics to inorganic products or low-molecular-weight organic fragments that can participate in the natural carbon cycle.
The range of Rhodococcus habitats is vast and diverse: these range from nutrient-rich (human and animal organisms, and plants) to oligotrophic (ground water, snow, and air) environments. Their adaptive traits include protective capsule-like structures and a lipophilic cell wall; colony dissociation and cellular polymorphism; temporarily dormant cyst-like cells and a low level of endogenous respiration (ensuring survival, e.g., upon prolonged starvation); production of carotenoids and extracellular glycolipids; nitrogen fixation in the presence of hydrocarbons; oligo- and psychrotrophy; acido-, alkalo-, halo-, xero-, thermo-, and osmotolerance; cell adhesion; and colonization of surfaces. Such a diversity of adaptive traits allows Rhodococcus accommodation in soil and aquatic environments, and possibly drove the appearance of epidemic variants in soil (water) and pathogenization of free-living forms. In extreme conditions of the polluted environment, rhodococci, as true saprophytic Gram-positive bacteria, are able to change their survival strategy in a manner that begins to exhibit traits of antagonism and pathogenicity [22][35].
2. Adaptive Cell Modifications of Rhodococci Exposed to Hydrocarbons and Other Environmental Pollutants
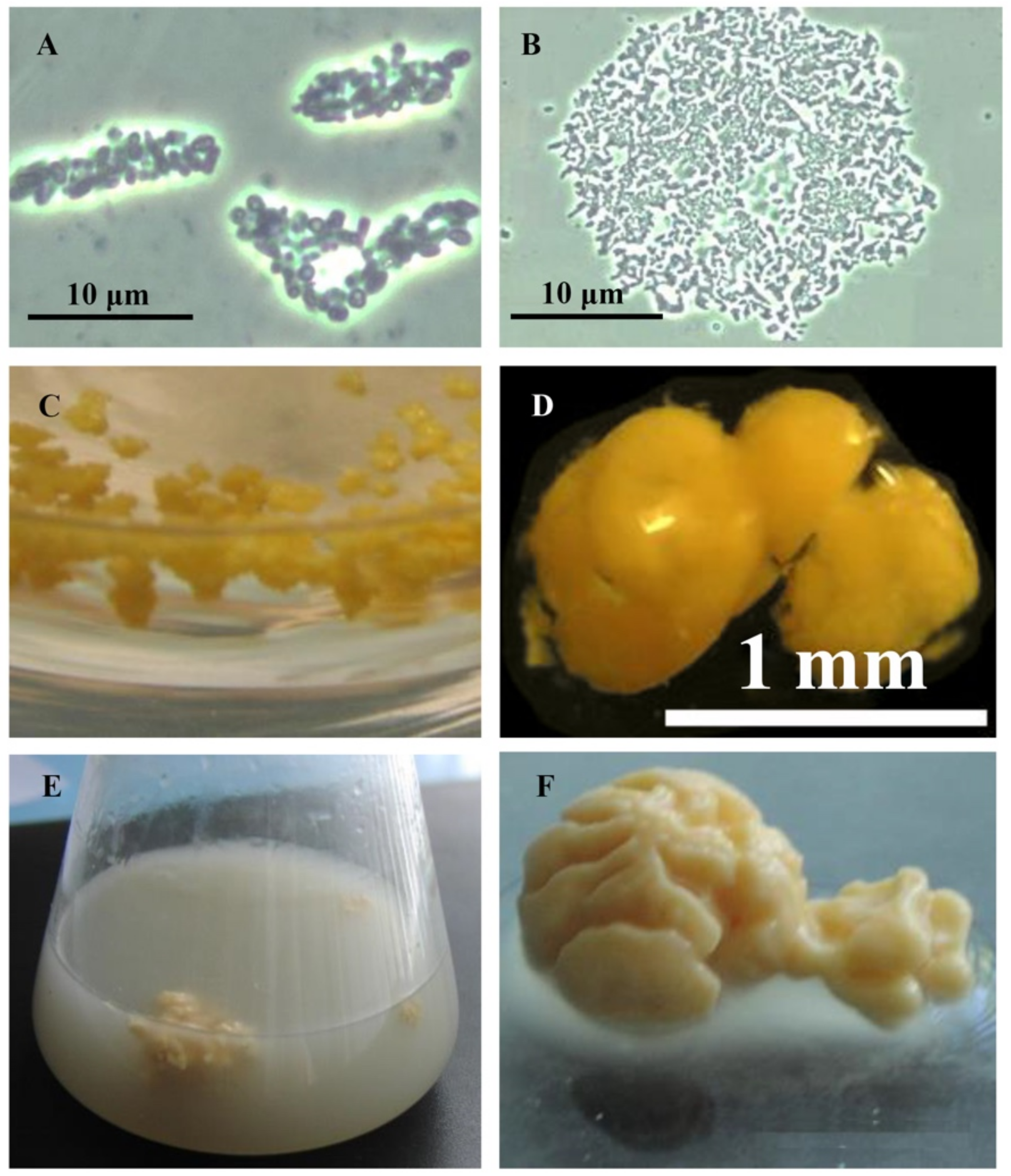
Rhodococci adapt to hydrophobic pollutants generally by forming separate multicellular aggregates (Figure 2). For example, when rhodococci were grown in the presence of liquid n-alkanes, planktonic microsized (25–70 µm in diameter) cell aggregates (microcolonies) of an elongated or rounded shape consisting of viable cells were formed during the first day (Figure 2A,B). On days 3–4, macroscopic compact multicellular (biofilm) formations (Figure 2C), or mucosal floccular strands (flocks) up to 1 cm in size diffusely located throughout the entire volume of the medium were observed. The spontaneous formation of biofilm clusters (cell aggregates, microcolonies) in the form of dense granules (up to 5 mm in diameter) seen by the naked eye, floating freely in the medium, and remaining until the end of the experiment, reduced the number of suspended cells in the culture broth (Figure 2D). By further self-immobilization, larger mushroom-like aggregates with a uniform structure passively floating in the liquid nutrient medium were formed (Figure 2E,F). In these cell clusters, the spatial arrangement of cells usually follows the rule of minimum diffusion distance, which ensures the fastest mass transfer between cells. Adhesion forces act at a distance of <20 nm [39].
3. Conclusions
Our goal is to highlight the structural, physiological, and biochemical features of a metabolically versatile group of nocardioform bacteria—representatives of the genus Rhodococcus , which are well-known biodegraders of hydrocarbons and other lipophilic organic compounds. These bacteria occupy one of the dominant positions in anthropogenically disturbed biotopes, and participate in their attenuation and restoration. The relative simplicity of the Rhodococcus cell structures is in harmony with the amazing perfection of their biological organization, and with their ability to form peculiar cellular adaptations having deep impacts primarily on cytology and physiology of rhodococci.
These materials substantiate the idea that the adaptive reactions of rhodococci to the negative effects of ecotoxicants are complex. In the presence of ecotoxicants, the developing Rhodococcus population changes smoothly towards the formation of more stable forms adapted to new stressful conditions. In this state, Rhodococcus cells are characterized by significant universal morphological changes. The discovered natural means of maintaining constant intracellular conditions in rhodococci exposed to various pollutants are considered to be mechanisms of their adaptation to the changing environment. They provide an improved tolerance of rhodococci to xenobiotics and the ability to change their survival strategy, and show signs of pathogenicity. The high competitiveness and survival of Rhodococcus species in any environment, in addition to their pathogenic potential, are determined by the ability of rhodococci to adhere to, aggregate, and colonize surfaces, and to change their lifestyle (from unicellular to multicellular, and from saprotrophic biodegraders to plant pathogens and animal intracellular parasites); the long-term persistence in unfavorable conditions; diauxotrophy; the formation of cyst-like cells and capsule-like structures; the enhanced cell surface hydrophobicity; ubiquity; and the antagonistic activities of some rhodococci. In medicine, the adaptive mechanisms are commonly referred to as “pathogenicity factors”, with adhesins and adhesion being particularly important as the mechanisms that trigger an infectious process.
Amid the pathogenization of saprotrophs under conditions of man-made pollution, the range of potentially dangerous microorganisms with “unprofessional” parasitism is expanding. There is a certain blurring of boundaries between pathogens and non-pathogens, and the idea of “universal” pathogenic factors of microorganisms is becoming more established. Moreover, it becomes increasingly difficult to predict which group of today′s saprophytes will join the list of pathogenic agents of disease tomorrow. At present, there is only an accumulation of research data on Rhodococcus survival strategies. The mechanisms of possible pathogenization of saprotrophic rhodococci have not been studied at the level of genome functioning and regulation of their metabolism. This has yet to be undertaken using modern genomic and post-genomic technologies with the use of system analysis.
This entry is adapted from the peer-reviewed paper 10.3390/pathogens10080974
References
- Goodfellow, M. Phylum XXVI. Actinobacteria phyl. nov. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Goodfellow, M., Kämpfer, P., Busse, H.-J., Trujillo, M.E., Suzuki, K., Ludwig, W., Whitman, W.B., Eds.; Springer: New York, NY, USA, 2012; Volume 5, pp. 33–34.
- Jones, A.L.; Goodfellow, M. Rhodococcus (Zopf 1891) emend. Goodfellow, Alderson and Chun 1998a. In Bergey’s Manual of Systematics of Archaea and Bacteria, Digital ed.; Trujillo, M.E., Dedysh, S., DeVos, P., Hedlund, B., Kämpfer, P., Rainey, F.A., Whitman, W.B., Eds.; John Wiley & Sons: New York, NY, USA, 2015; pp. 1–50.
- Gupta, R.S. Commentary: Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol. 2019, 10, 206.
- Busch, H.; Hagedoorn, P.-L.; Hanefeld, U. Rhodococcus as a versatile biocatalyst in organic synthesis. Int. J. Mol. Sci. 2019, 20, 4787.
- Ivshina, I.B.; Tyumina, E.A.; Kuzmina, M.V.; Vikhareva, E.V. Features of diclofenac biodegradation by Rhodococcus ruber IEGM 346. Sci. Rep. 2019, 9, 9159.
- Garrido-Sanz, D.; Redondo-Nieto, M.; Martín, M.; Rivilla, R. Comparative genomics of the Rhodococcus genus shows wide distribution of biodegradation traits. Microorganisms 2020, 8, 774.
- Alvarez, H.M.; Silva, R.A.; Cesari, A.C.; Zamit, A.L.; Peressutti, S.R.; Reichelt, R.; Keller, U.; Malkus, U.; Rasch, C.; Maskow, T.; et al. Physiolosgical and morphological responses of the soil bacterium Rhodococcus opacus strain PD630 to water stress. FEMS Microbiol. Ecol. 2004, 50, 75–86.
- LeBlanc, J.C.; Gonçalves, E.R.; Mohn, W.W. Global response to desiccation stress in the soil actinomycete Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 2008, 74, 2627–2636.
- Fanget, N.V.; Foley, S. Starvation/stationary-phase survival of Rhodococcus 1454 erythropolis SQ1: A physiological and genetic analysis. Arch. Microbiol. 2011, 193, 1–13.
- Corno, G.; Coci, M.; Giardina, M.; Plechuk, S.; Campanile, F.; Stefani, S. Antibiotics promote aggregation within aquatic bacterial communities. Front. Microbiol. 2014, 5, 297.
- Su, X.; Sun, F.; Wang, Y.; Hashmi, M.Z.; Guo, L.; Ding, L.; Shen, C. Identification, characterization and molecular analysis of the viable but nonculturable Rhodococcus biphenylivorans. Sci. Rep. 2015, 5, 18590.
- Röttig, A.; Hauschild, P.; Madkour, M.H.; Al-Ansari, A.M.; Almakishah, N.H.; Steinbüchel, A. Analysis and optimization of triacylglycerol synthesis in novel oleaginous Rhodococcus and Streptomyces strains isolated from desert soil. J. Biotechnol. 2016, 225, 48–56.
- Zhang, C.; Yang, L.; Ding, Y.; Wang, Y.; Lan, L.; Ma, Q.; Chi, X.; Wei, P.; Zhao, Y.; Steinbüchel, A.; et al. Bacterial lipid droplets bind to DNA via an intermediary protein that enhances survival under stress. Nat. Commun. 2017, 8, 15979.
- Raymond-Bouchard, I.; Tremblay, J.; Altshuler, I.; Greer, C.W.; Whyte, L.G. Comparative transcriptomics of cold growth and adaptive features of a eury- and steno-psychrophile. Front. Microbiol. 2018, 9, 1565.
- Firrincieli, A.; Presentato, A.; Favoino, G.; Marabottini, R.; Allevato, E.; Stazi, S.-R.; Mugnozza, G.-S.; Harfouche, A.; Petruccioli, M.; Turner, R.J.; et al. Identification of resistance genes and response to arsenic in Rhodococcus aetherivorans BCP1. Front. Microbiol. 2019, 110, 888.
- Cappelletti, M.; Presentato, A.; Piacenza, E.; Firrincieli, A.; Turner, R.J.; Zannon, D. Biotechnology of Rhodococcus for the production of valuable compounds. Appl. Microbiol. Biotechnol. 2020, 104, 8567–8594.
- Hu, X.; Li, D.; Qiao, Y.; Song, Q.; Guan, Z.; Qiu, K.; Cao, J.; Huang, L. Salt tolerance mechanism of a hydrocarbon-degrading strain: Salt tolerance mediated by accumulated betaine in cells. J. Hazard. Mater. 2020, 392, 122326.
- Sundararaghavan, A.; Mukherjee, A.; Suraishkumar, G.K. Investigating the potential use of an oleaginous bacterium, Rhodococcus opacus PD630, for nano-TiO2 remediation. Environ. Sci. Pollut. Res. 2020, 27, 27394–27406.
- Wang, C.; Chen, Y.; Zhou, H.; Li, X.; Tan, Z. Adaptation mechanisms of Rhodococcus sp. CNS16 under different temperature gradients: Physiological and transcriptome. Chemosphere 2020, 238, 124571.
- Pátek, M.; Grulich, M.; Nešvera, J. Stress response in Rhodococcus strains. Biotechnol. Adv. 2021, 107698.
- Goodfellow, M.; Jones, A.L.; Maldonado, L.A.; Salanitro, J. Rhodococcus aetherivorans sp. nov., a new species that contains methyl t-butyl ether-degrading actinomycetes. Syst. Appl. Microbiol. 2004, 27, 61–65.
- de Carvalho, C.C.C.R.; Costa, S.S.; Fernandes, P.; Couto, I.; Viveiros, M. Membrane transport systems and the biodegradation potential and pathogenicity of genus Rhodococcus. Front. Physiol. 2014, 5, 133.
- Ivshina, I.B.; Kuyukina, M.S.; Krivoruchko, A.V. Hydrocarbon-oxidizing bacteria and their potential in eco-biotechnology and bioremediation. In Microbial Resources: From Functional Existence in Nature to Industrial Applications; Kurtböke, I., Ed.; Elsevier: New York, NY, USA, 2017; pp. 121–148.
- Cappelletti, M.; Fedi, S.; Zannoni, D. Degradation of alkanes in Rhodococcus. In Biology of Rhodococcus. Microbiology Monographs, 2nd ed.; Alvarez, H.M., Ed.; Springer Nature: Cham, Switzerland, 2019; Volume 16, pp. 137–171.
- Acosta-González, A.; Martirani-von Abercron, S.-M.; Rosselló-Móra, R.; Wittich, R.M.; Marqués, S. The effect of oil spills on the bacterial diversity and catabolic function in coastal sediments: A case study on the Prestige oil spill. Environ. Sci. Pollut. Res. 2016, 22, 15200–15214.
- Kuyukina, M.S.; Ivshina, I.B. Bioremediation of contaminated environments using Rhodococcus. In Biology of Rhodococcus. Microbiology Monographs, 2nd ed.; Alvarez, H.M., Ed.; Springer Nature: Cham, Switzerland, 2019; Volume 16, pp. 231–270.
- Voronina, E.; Balandina, A.; Frolova, N.; Dubrovina, S.; Loginova, T. Effect of benzo(a)pyrene on the number of soil microorganisms of the genus Pseudomonas and Rhodococcus. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2021; Volume 723, p. 042010.
- Bej, A.K. Cold-tolerant alkane-degrading Rhodococcus species from Antarctica. Pol. Biol. 2000, 23, 100–105.
- de Carvalho, C.C.C.R. Adaptation of Rhodococcus erythropolis cells for growth and bioremediation under extreme conditions. Res. Microbiol. 2012, 163, 125–136.
- Goordial, J.; Raymond-Bouchard, I.; Zolotarov, Y.; de Bethencourt, L.; Ronholm, J.; Shapiro, N.; Woyke, T.; Stromvik, M.; Greer, C.W.; Bakermans, C.; et al. Cold adaptive traits revealed by comparative genomic analysis of the eurypsychrophile Rhodococcus sp. JG3 isolated from high elevation McMurdo Dry Valley permafrost, Antarctica. FEMS Microbiol. Ecol. 2016, 92, fiv154.
- Ivshina, I.B. Current situation and challenges of specialized microbial resource centres in Russia. Microbiology 2012, 81, 509–516.
- Catalogue of Strains of Regional Specialised Collection of Alkanotrophic Microorganisms. Available online: http://www.iegmcol/strains/index.html (accessed on 17 May 2021).
- Cuello, O.H.; Caorlin, M.J.; Reviglio, V.E.; Carvajal, L.; Juarez, C.P.; de Guerra, E.P.; Luna, J.D. Rhodococcus globerulus keratitis after laser in situ keratomileusis. J. Cat. Refr. Surg. 2002, 28, 2235–2237.
- Jones, A.L. Rhodococcus gordoniae sp. nov., an actinomycete isolated from clinical material and phenol-contaminated soil. Inter. J. Syst. Evol. Microbiol. 2004, 54, 407–411.
- Letek, M.; Gonzalez, P.; MacArthur, I.; Rodriguez, H.; Freeman, T.C.; Valero-Rello, A.; Blonco, M.; Buckley, T.; Cherevach, I.; Fahey, R.; et al. The genome of a pathogenic Rhodococcus: Cooptive virulence underpinned by key gene acquisitions. PLoS Genet. 2010, 6, e1001145.
- Ng, S.; King, C.S.; Hang, J.; Clifford, R.; Lesho, E.P.; Kuschner, R.A.; Cox, E.D.; Stelsel, R.; Mody, R. Severe cavitary pneumonia caused by a non-egui Rhodococcus species in an immunocompetent patient. Respir. Care 2013, 58, 47–50.
- Anastasi, E.; MacArthur, I.; Scortti, M.; Alvarez, S.; Giguère, S.; Vázquez-Boland, J.A. Pangenome and phylogenomic analysis of the pathogenic actinobacterium Rhodococcus equi. GBE 2016, 8, 3140–3148.
- Rahdar, H.A.; Mahmoudi, S.; Bahador, A.; Ghiasvand, F.; Heravi, F.S.; Feizabadi, M.M. Molecular identification and antibiotic resistance pattern of actinomycetes isolates among immunocompromised patients in Iran, emerging of new infections. Sci. Rep. 2021, 11, 10745.
- Bos, R.; van der Mei, H.C.; Busscher, H.J. Physico-chemistry of initial microbial adhesive interactions—Its mechanisms and methods for study. FEMS Microbiol. Rev. 1999, 23, 179–230.
- Matz, C.; Kjelleberg, S. Off the hook—How bacteria survive protozoan grazing. Trends Microbiol. 2005, 13, 909–915.
- Lekfeldt, J.D.S.; Rønn, R. A common soil fagellate (Cercomonas sp.) grows slowly when feeding on the bacterium Rhodococcus fascians in isolation, but does not discriminate against it in a mixed culture with Sphingopyxis witfariensis. FEMS Microbiol. Ecol. 2008, 65, 113–124.
- Corno, G.; Villiger, J.; Pernthaler, J. Coaggregation in a microbial predator–prey system affects competition and trophic transfer efficiency. Ecology. 2013, 94, 870–881.
- Korshunova, I.O.; Pistsova, O.N.; Kuyukina, M.S.; Ivshina, I.B. The effect of organic solvents on the viability and morphofunctional properties of Rhodococcus. Appl. Biochem. Microbiol. 2016, 52, 43–50.
- Rubtsova, E.V.; Kuyukina, M.S.; Ivshina, I.B. Effect of cultivation conditions on the adhesive activity of rhodococci towards n-hexadecane. Appl. Biochem. Microbiol. 2012, 48, 452–459.
- Denich, T.J.; Beaudette, L.A.; Lee, H.; Trevors, J.T. Effect of selected environmental and physico-chemical factors on bacterial cytoplasmic membranes. J. Microbiol. Methods. 2003, 52, 149–182.
- de Carvalho, C.C.C.R. Adaptation of Rhodococcus to organic solvents. In Biology of Rhodococcus. Microbiology Monographs, 2nd ed.; Alvarez, H.M., Ed.; Springer Nature: Cham, Switzerland, 2019; Volume 16, pp. 103–135.
- de Carvalho, C.C.C.R.; Fischer, M.A.; Kirsten, S.; Wurz, B.; Wick, L.Y.; Heipieper, H.J. Adaptive response of Rhodococcus opacus PWD4 to salt and phenolic stress on the level of mycolic acids. AMB Express 2016, 6, 66.
- Henson, W.R.; Hsu, F.F.; Dantas, G.; Moon, T.S.; Foston, M. Lipid metabolism of 1480 phenol-tolerant Rhodococcus opacus strains for lignin bioconversion. Biotechnol. Biofuels 2018, 11, 339.
- Su, X.; Guo, L.; Ding, L.; Qu, K.; Shen, C. Induction of viable but nonculturable state in Rhodococcus and transcriptome analysis using RNA-seq. PLoS ONE 2016, 11, e0147593.
- Urbano, S.B.; Albarracín, V.H.; Ordoñez, O.F.; Farías, M.E.; Alvarez, H.M. Lipid storage in high-altitude Andean Lakes extremophiles and its mobilization under stress conditions in Rhodococcus sp. A5, a UV-resistant actinobacterium. Extremophiles 2013, 17, 217–227.
- Kuyukina, M.S.; Ivshina, I.B. Production of trehalolipid biosurfactants by Rhodococcus. In Biology of Rhodococcus. Microbiology Monographs, 2nd ed.; Alvarez, H.M., Ed.; Springer Nature: Cham, Switzerland, 2019; Volume 16, pp. 271–298.
- Alvarez, H.; Steinbüchel, A. Biology of triacylglycerol accumulation by Rhodococcus. In Biology of Rhodococcus. Microbiology Monographs, 2nd ed.; Alvarez, H.M., Ed.; Springer Nature: Cham, Switzerland, 2019; Volume 16, pp. 299–332.
- Kalakoutskii, L.V. In the flow of time. In Proceedings of the IV International Conference “Microbial Diversity: Resource Potential”, Moscow, Russia, 23–25 November 2016; Ivshina, I.B., Kuyukina, M.S., Kamenskikh, T.N., Alfimova, L.A., Eds.; Institute of Ecology and Genetics of Microorganisms, Ural Branch, Perm State University, Russian Academy of Sciences: Moscow, Russia; Perm, Russia, 2016; pp. 121–122.
