Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Genetics & Heredity
In the present study, we investigate the widely expressed lncRNA LINC00493. We determine the structure of the LINC00493 transcript, its cell localization and influence on cell physiology. Our data demonstrate that LINC00493 has an influence on cell viability in a cell-type-specific manner. Furthermore, it was recently shown that LINC00493 has a sORF that is translated into small protein SMIM26. The results of our knockdown and overexpression experiments suggest that both LINC00493/SMIM26 transcript and protein affect cell viability, but in the opposite manner.
- LINC00493
- SMIM26
- long noncoding RNA
- lncRNA
- sORF
- MTT
- wound healing
1. Introduction
Advances in sequencing techniques revealed the transcription of non-coding regions of the genome which correspond to different groups, such as long noncoding RNA genes, small noncoding RNA genes, pseudogenes and immunoglobulin/T-cell receptor gene segments. Long noncoding RNAs (lncRNAs) are transcripts with lengths more than 200 nucleotides that are not translated into functional proteins. According to the GENCODE project (release 38) [1], the human genome contains 17,944 lncRNA genes. Data from the FANTOM CAT project revealed 27,919 human lncRNA loci [2]. The number of lncRNA genes is comparable to the number of protein-coding genes (19,954). However, lncRNAs make up only 0.03–0.20% of total RNA mass in the cell, whereas mRNAs make up 3–7% of it [3].
Initially thought to be transcriptional noise, several lncRNAs were discovered to be involved in gene expression regulation processes and affect cellular functions [4]. LncRNAs realize their functions through different intermolecular interactions: formation of a DNA–lncRNA triplex, formation of an lncRNA–RNA duplex and formation of an lncRNA–protein or lncRNA–chromatin complex [5]. These complexes may affect gene expression at the transcriptional [6,7,8,9] or post-transcriptional levels [10,11,12] and thus affect cellular phenotype.
Many lncRNAs are expressed in a tissue-specific manner, and their effect can vary in different cell types [13]. Moreover, lncRNAs’ expression can be altered in different pathological conditions, and their dysregulation may play an important role in disease progression [14]. To date, the lncRNADisease2.0 database contains entries about 19,166 lncRNAs associated with 529 diseases [15], including heart failure, cerebral injury, hypertension, acute kidney injury and cancer [16,17,18]. Genome-wide association studies (GWAS) indicate that lncRNA genes are enriched for trait- or disease-linked polymorphisms. Over 90% of all GWAS hits lie outside of known coding genes [19,20].
LncRNAs have an average length of about 3 kb and could contain up to 120 small open reading frames (sORFs) with a median of six sORFs per lncRNA. Recent studies proved that about 10,000 lncRNA genes in the mammalian genome contain sORFs less than 300 nt in length [21,22,23]. These sORFs could be translated into short peptides with key biological functions [24]. The presence of small peptides encoded by lncRNAs suggests that in some cases lncRNAs may have a dual function, or that the observed biological effect is contributed by the small protein, which means that this class of genes should be reclassified as protein coding [25,26,27].
In the present study, we investigate the widely expressed lncRNA LINC00493. We determine the structure of the LINC00493 transcript, its localization, protein-coding potential and its influence on cell physiology. Our data demonstrate the cell-type-specific role of LINC00493. During our work, LINC00493 was predicted to contain a sORF that could translate a small protein—SMIM26. We collected and described all the existing data on this protein. Using knockdown and overexpression experiments, we obtain data suggesting that both LINC00493/SMIM26 transcript and protein affect cell viability, but in the opposite manner.
2. Investigation of LINC00493/SMIM26 Gene Suggests Its Dual Functioning at mRNA and Protein Level
2.1. LINC00493 Transcript Structure
The LINC00493 gene was predicted through the ENCODE project. The UCSC Genome Browser [28] shows that LINC00493 is located on human chromosome 20p11.23 and consists of two or three exons, according to mRNA and EST sequence data from RefSeq and Ensembl databases. In contrast to the protein-coding genes, the lncRNA gene annotations tend to have poorly defined boundaries, because of weak conservation, low and tissue-specific expression and lack of characteristic hallmarks of transcription initiation and termination [29]. Therefore, to define the exact structure of the LINC00493 transcript we performed reverse transcription (RT)-PCR and rapid amplification of cDNA ends (RACE) analysis on total RNA from HEK293T, HeLa cell lines and human primary skin fibroblasts. RT-PCR analysis revealed that the LINC00493 transcript consists of two exons and RACE showed the exact 5′ and 3′ cDNA ends (Figure 1A).
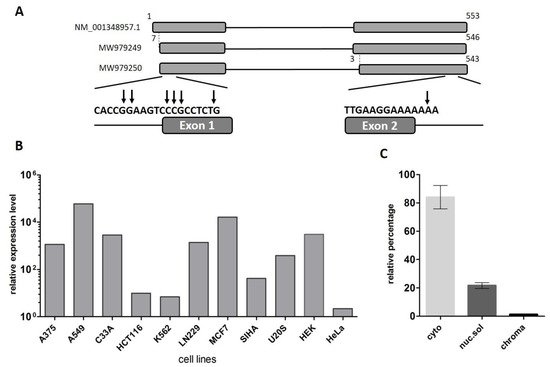
Figure 1. Analysis of the structure and expression of the LINC00493 transcript. (A) Scheme of the LINC00493 gene (according to RefSeq NM_001348957.1 sequence). Results of 5′ and 3′ RACE analysis are presented under the scheme. The vertical arrows represent the genomic position of exact 5′ and 3′ ends. Nucleotide numbering was based on reference sequence NM_001348957.1 (B) Relative expression of LINC00493 transcript in 11 human cell lines was detected by RT-qPCR. (C) Subcellular localization of LINC00493 was detected by qPCR of RNA isolated from cytoplasmic (cyto), nuclear-soluble (nuc.sol) and chromatin-bound (chroma) fractions of HEK293T cells. The error bars represent SEM (standard error mean).
According to the Ensembl database, there are two LINC00493 isoforms, ENST00000411646.1 and ENST00000435844.3, which differ by three nucleotides at the beginning of the second exon. Our RT-PCR analysis confirmed that LINC00493 has two isoforms, expressed in all analyzed cell lines. The total length of the short and long isoforms was 500 and 497 bp, respectively. The nucleotide sequences of short and long isoforms were deposited into GenBank under accession numbers MW979249 and MW979250. The difference in the sequencing signal suggests that the major long isoform is expressed at an approximately 3-fold higher level than the minor one.
2.2. LINC00493 Is Widely Expressed in Human Tissues and Cell Lines
To identify LINC00493 expression profile, we provided analysis of the FANTOM5 and GTEx expression data. We observed that LINC00493 is highly expressed in most human cell lines and tissues. An expression profile of LINC00493 in 889 human samples from FANTOM5 classified this gene as a housekeeping gene [30]. We confirmed the high widespread expression level of this transcript using RT-qPCR analysis of 11 human cell lines, as well as human primary skin fibroblasts (Figure 1B). The highest expression level was observed in A549, MCF7 and HEK293T cell lines.
2.3. Cytoplasmic Localization of LINC00493
LncRNA subcellular localization is closely related to its biological function. Some lncRNAs play a role in a transcriptional regulation through their interaction with chromatin, while others are found in the cytoplasm and affect the post-transcriptional control of gene expression or could be translated into small peptides. We investigated the subcellular localization of the LINC00493 transcript using the soft lysis method. RNA was isolated from cytoplasmic, nuclear and chromatin-bound fractions of cells. To determine the level of the investigated transcript in each fraction we performed RT-qPCR. Our analysis revealed that the LINC00493 transcript is localized predominantly in cytoplasm (Figure 1C). This result suggests that the function of the LINC00493 transcript is not related to the transcription regulation and chromatin binding. The observed results highlight similarities between the investigated lncRNA and mRNAs, such as sequence length, high expression level and accumulation in the cytoplasm.
2.4. Knockdown of LINC00493 Affects Cell Growth in a Cell-Type-Specific Manner
To determine the function of LINC00493, we analyzed previously published CRISPRi-based data for functional long noncoding RNA loci in human cells [13] and discovered that LINC00493 modified cell growth in a cell-type-specific manner. To confirm the cell-type-specific role of LINC00493, we performed knockdown experiments using RNA interference in three human cell lines: HEK293T, A375 and MDA-MB-231.
LINC00493 knockdown efficiency was about 60–70% (Figure 2A). After knockdown, cell proliferation was measured by MTT assay, and cell migration was examined using wound-healing assay. We revealed that LINC00493 knockdown reduced cell viability in HEK293T and A375 cell lines, while the opposite effect was observed in MDA-MB-231 (Figure 2C). Thus, knockdown experiments confirmed that downregulation of LINC00493 affected cell proliferation activity in a cell-type-specific manner. On the other hand, wound-healing assay revealed that LINC00493 knockdown did not affect cell migration (Figure 2B,D).
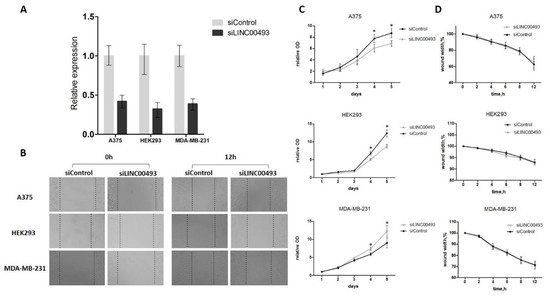
Figure 2. LINC00493 knockdown has a cell-type-specific effect. (A) LINC00493 knockdown efficiency in A375, HEK293T and MDA-MB-231 cell lines. (B) The migration ability of A375, HEK293T and MDA-MB-231 cells after transfection of siLINC00493 and siControl was measured by wound-healing assay. Representative images of wound-healing experiments are shown. (C) MTT assay reveals the effect of LINC00493 knockdown in 3 cell lines. Relative optical density value of A375, HEK293 and MDA-MB-231 cells with LINC00493 knockdown and control cells in the MTT assay. (D) Summary graph showing typical wound-healing rates by A375, HEK293 and MDA-MB-231 cells after LINC00493 knockdown. Error bars represent the mean ± SEM (standard error mean) of three independent experiments. * p < 0.01, vs. control (according to Mann–Whitney U test).
2.5. Small Protein Is Translated from LINC00493 RNA
According to the Human Protein Atlas [31], LINC00493 contains a small open reading frame that could be translated to a 94/95-amino-acid protein—SMIM26. Using GWIPS-viz [32] and Trips-Viz [33], which provide ribosome profiling data, we confirmed that LINC00493 has a strong ribosome association in the predicted sORF region (Figure 3C). The predicted secondary and tertiary structures of SMIM26, constructed using an improved predictor of protein structure [34], showed that the protein contains two alpha helixes; one of them is supposed to be a transmembrane domain (Figure 3B,D). A number of tools (Phobius, SPOCTOPUS, MEMPACK) predict that SMIM26 is localized in the membrane. Despite this fact, the Human Protein Atlas immunocytochemistry analysis revealed that this protein is localized mainly in the nucleoplasm and partially in the cytosol [31]. However, the function of the protein remains unknown. Comparative analysis of amino-acid and nucleic sequences of LINC00493 reveals that the gene is evolutionarily conserved among mammals both at the RNA and protein level (Figure 3A). This fact supports a potential functional role for SMIM26, while the influence of this protein on cell migration and viability was not shown previously.
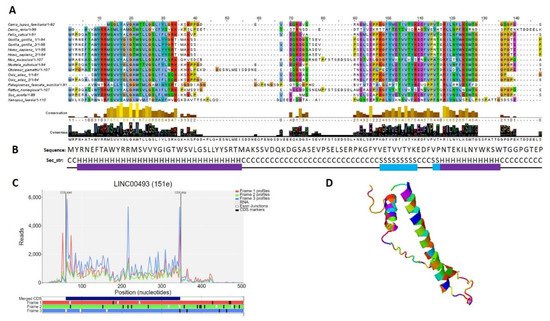
Figure 3. Analysis of the structure of the SMIM 26. (A) Multiple-species alignment of the amino acid sequences of the LINC00493-encoded small protein SMIM26. (B) Predicted secondary structure (H: helix; S: strand; C: coil) of SMIM26. (C) Distribution of the RPF reads in the LINC00493-ORF. (D) Predicted 3D structure of SMIM26.
2.6. SMIM26 Protein Affects Cell Viability
Data from knockdown experiments revealed that the LINC00493 gene is important for cell viability. However, it remains unclear whether the observed effect is associated with small protein or with the RNA itself. To investigate separately the effect of SMIM26 protein and the LINC00493 transcript on cell viability, we cloned a full-length LINC00493 cDNA into pcDNA3.1-GFP vector (pcDNA3.1) and obtained LINC00493_WT plasmid. Next, we mutated the SMIM26 start-codon by site-directed mutagenesis to prevent the translation of small protein and obtained the LINC00493_MUT plasmid. Wild-type and mutant constructs, along with an empty pcDNA3.1, were transfected into A375 and HEK293T cells and cell viability was measured using the MTT test. The expression of LINC00493 was increased about a thousand-fold times by qPCR (Figure 4A).
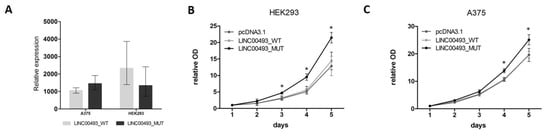
Figure 4. Influence of LINC00493/SMIM26 overexpression on cell viability. (A) LINC00493_WT and LINC00493_MUT overexpression efficiency in A375 and HEK293 cell lines. (B,C) Relative optical density value of HEK293 and A375 cells treated by LINC00493_WT, LINC00493_MUT and control cells in the MTT assay. Cells with overexpression of mutated LINC00493 showed higher viability than those transfected with WT LINC00493 or empty pcDNA3.1 vectors. Error bars represent the mean ± SEM (standard error mean) of three independent experiments. * p < 0.01, vs. control (according to Mann–Whitney U test).
Interestingly, overexpression of WT LINC00493 did not influence the number of cells, while overexpression of LINC00493 with a mutated start-codon demonstrated increased cell viability (Figure 4B,C). This observation suggests that, even in the absence of SMIM26 protein translation, the LINC00493 transcript itself has an effect on cell viability. However, the difference between WT and MUT LINC00493 overexpression indicates that SMIM26 protein could also affect cell viability, but in the opposite manner.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22168477
This entry is offline, you can click here to edit this entry!
