The identification of the molecular mechanisms controlling early cell fate decisions in mammals is of paramount importance as the ability to determine specific lineage differentiation represents a significant opportunity for new therapies. Pancreatic Progenitor Cells (PPCs) constitute a regenerative reserve essential for the maintenance and regeneration of the pancreas. Besides, PPCs represent an excellent model for understanding pathological pancreatic cellular remodelling. Given the lack of valid markers of early endoderm, the identification of new ones is of fundamental importance. Both products of the Ink4a/Arf locus, in addition to being critical cell-cycle regulators, appear to be involved in several disease pathologies. Moreover, the locus’ expression is epigenetically regulated in ES reprogramming processes, thus constituting the ideal candidates to modulate PPCs homeostasis.
1. Introduction
Alterations in PPCs (Pancreatic Progenitor Cells) homeostasis are fundamental to physiological processes, such as organ development and pathological events. In particular, PPCs are essential for the maintenance and regeneration of organs and tissues that exhibit high rates of cell turnover or regenerative reserve. Interestingly, events associated with an early epithelial-to-mesenchymal transition (EMT) phenotype closely resemble PPCs cellular transition that occurs during normal pancreas embryogenesis
[1][2]. Experiments on genetically engineered mice revealed that depletion of progenitor cells leads to acute or delayed failure in tissue homeostasis, limited regenerative potential, and physiological aging
[3]. Besides physiological aging, senescence is a growth-arrest mechanism triggered by different insults to protect cells from hyper-proliferative signals and various forms of stress, representing an important barrier against tumor formation
[3].
The
CDKN2A locus encodes two tumor suppressors, p16INK4a (INK4a) and p14ARF (ARF), and is among the most frequently mutated in human cancers
[4]. Inactivation of Arf and Ink4a in mice induces tumors with complete penetrance, while in several human cancers, both epigenetic silencing and mutational inactivation of these genes have been described
[5]. Interestingly, the locus is found mutated or epigenetically silenced in 30–70% of PanINs (pancreatic intraepithelial neoplasms), a percentage that becomes as high as 95% in full-blown tumors
[6]. Asides from being involved in cancer, both INK4a and ARF are involved in differentiation, apoptosis, and senescence in a number of cellular contexts
[7][8]. Interestingly, both INK4a and ARF are expressed during pancreas organogenesis and are tightly correlated with PPCs’ quiescence
[9]. Several lines of evidence suggested that increased transcription of INK4A and ARF provokes senescence in various cell types with a causative role in age-associated degenerative diseases. Hence, the comprehension of the mechanisms regulating the expression of these genes has major implications for both cancer and degenerative disorders. In beta cells, it is known that the lack of INK4a expression leads to the overexpression of CDK4 and consequently to an increase in cell proliferation, rising insulin secretion, and causes pancreatic hyperplasia
[9]. Germline
CDKN2A mutations have also been occasionally found in pancreatic cancer patients and families without familial melanoma, in which the locus involvement has been described in several systems
[10][11][12]. Variants of
CDKN2A/B are involved in susceptibility to type 2 diabetes highlighting its possible role in Beta-cell function and regeneration
[13]. Besides their function as a barrier to tumor progression, a number of studies demonstrated that both proteins are implicated in developmental processes
[7].
In this study, we followed the Ink4a/Arf locus expression during pancreatic commitment in mouse ESC. We provide data showing that upon mESCs differentiation in vitro, Ink4a/Arf expression sharply increases both at a transcriptional and protein level. In particular, both ARF and INK4a are expressed during the early stages of murine endoderm specification in a subset of cells marked by the expression of the Nepn (Nephrocan) gene at E7.5–11.5 of mouse development. Furthermore, mESCs treatment with 5-aza-2-deoxyazacytidine (AZA) and trichostatin (TSA), in addition to allowing the expression of PPC markers, also induces an increase in Ink4a/Arf gene expression, thus suggesting that an epigenetic-dependent mechanism controls Cdkn2a locus remodelling activated during PPCs differentiation.
2. Ink4a/Arf Gene Expression Increases in mESCs Induced to Differentiate towards Pancreatic Lineage
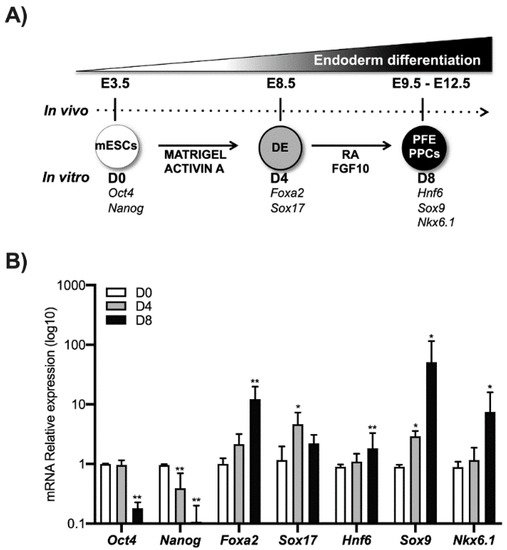
Using an established differentiation protocol
[14], E14 mouse ESCs were induced to differentiate into pancreatic progenitors. In particular, mESCs cells expressing
Oct4 and
Nanog marked as D0 were grown in matrigel and activin A until the expression of the DE markers Foxa2 and Sox17 (typically 4 days, indicated herein as D4). These cells were further treated for 4 days with RA and FGF10 to induce the expression of molecular markers such as Hnf6, Sox9, and Nkx6.1 that are typical of the PFE. These cells are highly enriched in PPCs (herein indicated as D8) and represent a good in vitro model to investigate the molecular mechanisms that occur during pancreatic differentiation (
Figure 1).
Figure 1. ESCs differentiation toward pancreatic lineage. (A) ESCs (D0) were cultured for 4 days treated with activin A (30 ng/mL) (D4), the definitive endoderm (DE) cells were cultured for another 4 days in the presence of both RA and FGF10 to obtain the Posterior Foregut Endoderm (PFE) and enrichment of PPCs (D8). The correspondence of the in vivo and in vitro phases was indicated. (B) Pluripotency markers (Nanog and Oct4), definitive endoderm markers (Foxa2 and Sox17), Posterior Foregut (Hnf6), and Pancreatic Progenitor Cell markers (Sox9 and Nkx6.1) were analyzed by qPCR. Data points represent the average of three independent experiments ± SD. * p < 0.05; ** p < 0.01.
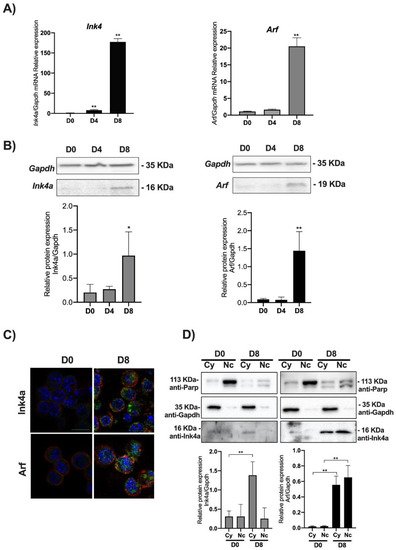
We thus analyzed the level of expression of the Ink4a/Arf locus’ products evaluating both the mRNA and protein levels of Ink4a and Arf by qPCR and Western blot. Transcriptional analysis of Ink4a and Arf genes showed that their expression increases as long as the cell’s differentiation proceeds toward the PPC’s lineage. The Ink4a gene expression significantly increased at DE (D4) by around 20 times compared to the control (D0), while in PFE (D8), its transcriptional expression was increased by nearly 200 times compared to the control (Figure 2A).
Figure 2. Ink4a and Arf expression increase in PPCs. ESCs (D0) were induced to differentiate towards PPCs (D4 and D8) and Ink4a and Arf gene expression analyzed by qPCR, Western blot, and immunofluorescence staining. (A) Relative Ink4a and Arf gene expression vs. Gapdh is reported as folds increase. (B) Western blots and densitometric analysis of Ink4a and Arf protein levels relative to Gapdh. (C) Merged images of the immunofluorescence staining at D0 and D8 performed with anti-Ink4a and Arf antibodies (green), rhodamine-phalloidin (for F-actin, red), and DAPI (for DNA, blue). Images were taken with a Zeiss confocal laser-scanning microscope LSM 510 (Oberkochen, Germany), scale bar, 15 μm. (D) Subcellular fractionation of ESCs at D0 and D8: Western blot analysis of equal amounts of cytoplasmic (30 μg) and nuclear (30 μg) protein extracts. The efficiency of cellular fractionation was checked with anti-Parp (for nuclei) and anti-Gapdh (for cytoplasm) antibodies. Normalized Ink4a and Arf band intensities are expressed relative to Gapdh expression in each experimental point. The data are expressed as average of three independent experiments ± SD, * p < 0.05; **, p < 0.01.
Arf gene expression mirrors the kinetics of
Ink4a. While remaining similar between D4 and D0, the transcript’s level is significantly increased by 20 times in D8. Quantification of Ink4a and Arf immunoblots showed a sharp increase of both proteins in D8 compared to D0 (see graphs in
Figure 2B). The immunofluorescence staining confirmed an increased expression of both proteins in D8 (
Figure 2C and
Figure S2). These results were further confirmed for both proteins by nuclear/cytoplasmic fractionation in D0 and D8 (
Figure 2D). Ink4a appeared poorly expressed in D0, acquiring a clear cytoplasmic localization in D8 (
Figure S1). On the other end, Arf showed a sharp increase both in the nucleus and cytoplasm in D8 (
Figure 2D and
Figure S3).
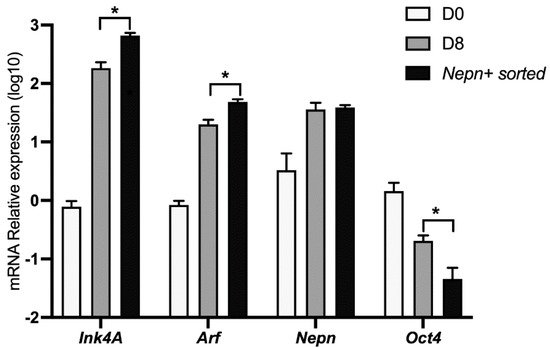
3. Ink4a/Arf Gene Expression Correlates with Endoderm-Nepn Cells
It has been shown that the
Nepn gene marks the early stages of murine endoderm specification and is required for correct endoderm differentiation
[15][14]. To further characterize the kinetics of the
Ink4a and
Arf expression in our cellular system, we differentiated ES cells as described before and analyzed the locus’ expression in a pure subpopulation of
Nepn expressing cells. Differentiating cells were sorted by FACS in order to obtain a population enriched in
Nepn expressing progenitor cells. When we analyzed
Cdkn2a’s transcript levels by qPCR, we found a significant enrichment of both
Ink4a and
Arf expression in Nepn-sorted cells. Moreover, a comparison of the expression level dynamics strongly suggests that the locus expression is confined in the Nepn+ cell state (
Figure 3).
Figure 3. Ink4a and Arf expression mark Nepn-expressing cells. mESCs were induced to differentiate for 8 days as described, and cells expressing Nepn-expressing cells were isolated by FACS. Total RNA was extracted and analyzed by qPCR to check Ink4a, Arf, Nepn, and Oct4 gene expression. Data points represent the average of independent experiments performed in triplicate ± SD. *, p < 0.05.
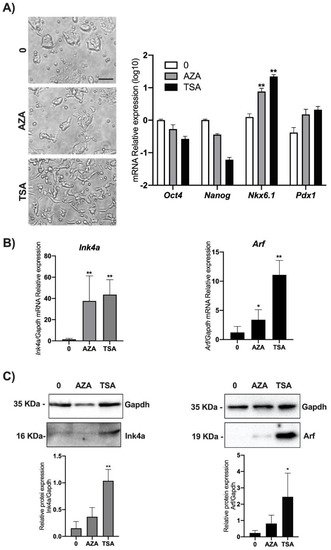
4. DNA Methylation and Chromatin Acetylation Affect Ink4a and Arf Gene Expression in ESCs
It is known that the
Cdkn2a locus is not expressed in ES and in iPS (induced Pluripotent Stem) cells but acquires the epigenetic marks of a bivalent chromatin domain, thus retaining the ability to be reactivated if required after differentiation onset
[16][17]. Epigenetic suppression of the INK4A-ARF locus has been widely shown, and greatly promotes survival and cloning efficiency in individualized human ESCs via deposition of the H3K27me3-mark to INK4A-ARF locus
[18][19]. In order to verify if epigenetic modifications, specifically DNA methylation and chromatin deacetylation, were responsible for the Ink4a and Arf regulation in ESCs, we performed treatments of cells with widely used epi-drugs. In particular, we used either the demethylating agent AZA or the HDAC inhibitor TSA. mESCs were treated with 25 µM AZA or with 100 nM TSA. Twenty-four hours post-treatment,
Ink4a and
Arf expression in mESCs cells was simultaneously analyzed by qPCR and Western blot. Before lysis, cells were observed by phase-contrast microscopy, and we noticed that cells treated with TSA, in contrast to AZA treated cells, acquired an elongated morphology (
Figure 4A left). The analysis of pluripotency markers such as
Oct4 and
Nanog [20][21] and differentiation markers such as
Nkx6.1 and
Pdx1 [22] are in line with the epigenetic pattern of regulation of these genes suggesting the priming of a differentiation program (
Figure 4A right).
Figure 4. Effect of AZA and TSA on Ink4A and Arf gene expression in mESCs. ESCs were treated either with 25 μM AZA or 100 nM TSA and with 0.1% DMSO as control (0). (A) After 24 h, treated and control mESCs were imaged using phase-contrast microscopy with a 10× objective (left) (scale bar, 60 μm). The experiment was performed in triplicate, and different fields were observed. Total RNA and proteins were isolated for each experimental point, and the levels of genes expression of Oct4, Nanog, Nkx6.1, and Pdx1 were analyzed as experimental control (right). Ink4a and Arf expression was analyzed by (B) qPCR and (C) Western blot assays. Data are shown as mean ± SD from three independent experiments, normalized with Gapdh, and reported with respect to the control (0). *, p < 0.05; **, p < 0.01.
While we found that both treatments significantly increase both Ink4a and Arf transcriptional levels, analysis by Western blot revealed a significant increase only upon TSA treatment (
Figure 4B,C and
Figure S4).
This entry is adapted from the peer-reviewed paper 10.3390/biom11081124