Resveratrol is a plant secondary metabolite known for its therapeutic applications as an antioxidant, anti-cancer, anti-inflammatory, anti-aging, cardio-protective, and neuroprotective agent. Topical formulas of resveratrol are also used for skin disease management and in cosmetic industries. Due to its importance, high resveratrol production is urgently required. Yeasts were proven to be excellent host candidates for resveratrol production.
- resveratrol
- yeast hosts
- pathway engineering
- metabolic engineering
1. Introduction
Resveratrol (3,4′,5-trihydroxystilbene) is a natural polyphenolic phytoalexin that belongs to the stilbenoids group of secondary metabolites [1][2][3]. Stilbenes are known for their ability to protect plants from UV light and the effect of chemical fertilizers [4], and for the defense of plants against biotic stresses such as bacterial, fungal, or nematode infections [3][5]. Resveratrol was first identified in 1940 as a constitutive compound of white hellebore roots (Veratrum grandiflorum) [6]. The richest source of resveratrol was found in Polygonum cuspidatum roots, the extract of which is widely used in traditional Chinese and Japanese medicine to treat human fungal diseases, such as gonococcal infection, suppurative dermatitis, tinea favosa and tinea pedis infections, hyperlipidemia, arteriosclerosis, and inflammations [1][7]. Thus far, resveratrol has been found in various plant species, such as grape berries [8], blueberries, cranberries (Vaccinium spp.) [9][10], blackberries, mulberries (Rubus and Morus spp.) [11][12], peanuts (Arachis hypogaea) [13], and jackfruit (Artocarpus heterophyllus) [11]. Naturally, there are two isomeric forms of resveratrol (Figure 1) but the trans isomer is the biologically active form [4]. However, under high pH values or UV-light exposure, resveratrol converts from the trans isoform into the cis isomer [14]. Both isomers exist as glucosides, and 3-O-β-d-resveratrol glucoside (piceid or polydatin) is the richest source of resveratrol in the glucoside form [1][14][15].
2. Resveratrol Biosynthesis in Nature
Stilbenes are plant secondary metabolites built from joining two aromatic rings with an ethylene bridge (1,2-diphenylethylene backbone) to form the basic C6-C2-C6 skeleton. Stilbenes share many similarities in their structure, biosynthesis, and biological activities with phenylpropanoids and flavonoids, which originate from the same pathway. The key components for biosynthesis of stilbenes are malonyl-CoA, phenylalanine, and tyrosine (Figure 2). In glycolysis pathway, glucose is converted into pyruvic acid, which leads to acetyl coenzyme A. The latter is the precursor of malonyl-CoA, which plays an essential role in biosynthesis pathways for many secondary plant compounds. The condensation of erythrose-4-phosphate (four carbons), which is generated from the pentose phosphate pathway (parallel reaction pathway to glycolysis) and phosphoenol pyruvate (three carbons, an intermediate in glycolysis), yields the seven-carbon shikimate skeleton. The latter compound is then used in the shikimic acid pathway to generate the aromatic amino acids phenylalanine and tyrosine [1][16].
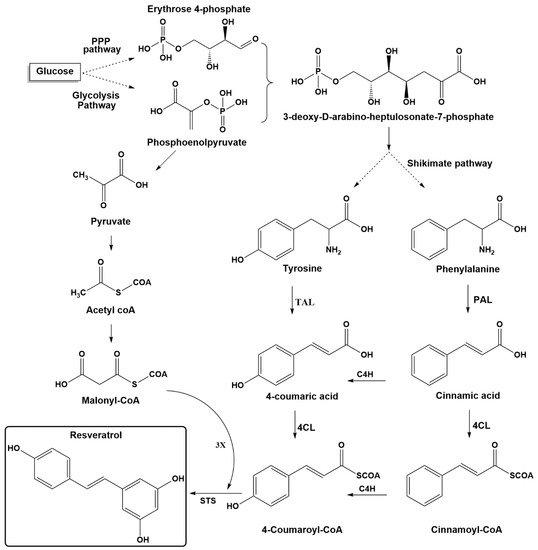
The biosynthesis pathway for resveratrol starts with phenylalanine or tyrosine, as shown in Figure 2. The first step is to obtain para-coumaric acid from both pathways. The phenylalanine ammonia-lyase (PAL) transforms phenylalanine into cinnamic acid, which is further processed to generate 4-coumaric acid (para-coumaric acid) using cinnamate-4-hydroxylase (C4H). Tyrosine ammonia-lyase (TAL) can directly generate p-coumaric acid from tyrosine. The second step includes transformation of para-coumaric acid into para-coumaroyl-CoA by para-coumaroyl coenzyme A ligase (4CL). The final step involves the condensation of three units of malonyl-CoA with p-coumaroyl-CoA through stilbene synthase (STS) [1][17][18]. Starting from resveratrol, more modifications can be accomplished on its structure by various decorating enzymes.
3. Resveratrol Production by Transgenic Yeasts
Microorganisms have been essential sources for producing pharmaceutically and industrially important compounds for decades due to low-cost cultures, fast production, the ability to construct purification processes, the ease of manipulating their genetic components, as well as the availability of protein and metabolic engineering tools [19]. As eukaryotic organisms, yeast species have the ability to express genes coding for plant-derived enzymes, such as cytochrome P450 enzymes; to perform post-translational modification (such as glycosylation); and to functionally fold the eukaryotic recombinant protein [20][21]. Additionally, yeasts and plants share similar intracellular compartments such as the endoplasmic reticulum, which supports eukaryotic and membrane proteins biosynthesis [22]. Another advantage of using yeasts is that Saccharomyces cerevisiae (the most-used yeast species for resveratrol production) is a food-grade organism that can be used safely in human nutrition and pharmaceutical products [23]. Similar to S. cerevisiae, Yarrowia lipolytica is classified as a generally recognized as safe organism (GRAS) [24] and both are strong candidates for use as cell factories for resveratrol production.
3.1. Pathway Engineering
The introduction of an entire biosynthetic pathway into the microorganism provides the ability to produce resveratrol from its precursors (L-phenylalanine or L-tyrosine) or low-cost materials such as glycerol, glucose, or ethanol. In this method, PAL or TAL, depending on the used pathway; C4H; 4CL; and STS encoding genes are transferred into the chosen host. [18][25][26]. One of the first attempts to entirely reconstruct the resveratrol pathway was conducted by Zhang et al. in 2006 in S. cerevisiae. TAL from Rhodobacter sphaeroides, 4CL from Arabidopsis thaliana, and STS from V. vinifera (4CL::STS fusion protein) were introduced into the S. cerevisiae strain WAT11. The expression of the coupled genes coding for the fusion protein increased the resveratrol titer up to 15-fold compared with the co-expression of the genes encoding the separate enzymes, which emphasized the importance of the fusion protein and the spatial localization of these two related enzymes to improve resveratrol production [27].
In a recent study, Y. lipolytica was engineered using TAL from Flavobacterium johnsoniae, PAL and STS from V. vinifera, and C4H and 4CL1 from A. thaliana. The strains showed the ability to produce resveratrol using both pathways separately (using tyrosine or phenylalanine as precursors) or in combination. The importance of gene overexpression by increasing the gene copy number in the resveratrol pathway was clearly demonstrated in this study, in which strains harboring two copies of the PAL, C4H, 4CL1, and STS genes or two copies of TAL, 4CL1, and STS exhibited higher performances than single-copy sets of genes. The best results were obtained from a strain containing two copies of PAL, C4H, TAL, 4CL1, and STS, which produced a high titer of resveratrol, reaching 450 mg/L under fermentation conditions using 100 g/L glycerol as the sole carbon source, which is the highest reported amount of resveratrol produced from the expression of only the resveratrol pathway [28].
Engineering partial pathways or selected genes is an alternative strategy to produce resveratrol. In this case, PAL, TAL, and C4H are usually excluded from the pathway construction, and para-coumaric acid is used as the precursor. S. cerevisiae FY23 was the first used for resveratrol pathway construction in yeast. The 4CL216 from a hybrid poplar and vst1 from a grape vine were constitutively expressed in the strain. After feeding with 5 mM p-coumaric acid, the recombinant strain produced 1.45 µg/L resveratrol [29].
Using synthetic scaffolds is another strategy used for improving resveratrol production. GTPase binding domain (GBD), Src homology 3 domain (SH3) with 4CL1 from A. thaliana, and PSD95/DlgA/Zo-1 domain (PDZ) with STS from V. vinifera were recruited and optimized in S. cerevisiae WAT11 cells. Using this scaffold strategy increased the resveratrol titer by 2.7-fold compared with the fusion enzyme strategy for the same genes [27][30]. This indicates the effectiveness of protein scaffolds in improving resveratrol synthesis and in increasing pathway enzyme activity compared with the protein fusions strategy.
The importance of selecting pathway genes and codon-optimizing them was explored in a recent report, in which six STS genes from different sources (PhStS, PcPKS5, MaSTS3, RtSTS, VvVST1, and AhSTS) were codon-optimized and then co-expressed with 4CL from Plagiochasma appendiculatum in S. cerevisiae W303. Differences in production time and final yield titer were observed among the different strains [31]. Codon optimization for resveratrol pathway genes has also been observed in several studies in both S. cerevisiae and Y. lipolytica [32][33][34][35][36]. The highest resveratrol titer, which was produced by the expression of partial pathway genes (4CL and STS) in yeasts, was obtained from industrial Brazilian sugar-cane-fermenting yeast, and the titer was 391 mg/L of resveratrol [37].
3.2. Host Metabolic Engineering (Non-Pathway Genes)
Metabolic engineering of microbial hosts for resveratrol production has achieved significant progress in recent years. The main remaining obstacles for microbial production of resveratrol are the precursor availability and the low activity of stilbene synthase in heterologous hosts. Hence, the primary strategies for increasing productivity using microbial cell factories are increasing the precursor supply (aromatic amino acids and malonyl-CoA) via genetic manipulation of the strain and improving the activity of key enzymes via protein engineering.
To increase the precursor levels, the introduction of non-pathway exogenous genes and pathway redirection are crucial. Overproduction of aromatic amino acids can be easily achieved by engineering the shikimate pathway. Naturally, the majority of malonyl-CoA is used in fatty acid biosynthesis, leaving a minimal level to be used in resveratrol biosynthesis. Thus, increasing this precursor in microbial hosts provides the opportunity for extra enhancement in resveratrol biosynthesis in the host platform.
Two main strategies are usually applied to enhance the malonyl-CoA pool. The first is to improve acetyl-CoA carboxylation into malonyl-CoA via the acetyl-CoA carboxylase (ACC) enzyme. The other strategy is to inhibit malonyl-CoA consumption by repressing the biosynthesis of fatty acids [38]. Since the blocking of malonyl-CoA consumption and direct knockouts of the fab genes were found to be lethal to microorganisms [39], the inhibition has been accomplished in bacterial hosts via three procedures: the addition of cerulenin antibiotic to inhibit FabB and FabF [40][41]; using antisense RNA to repress the fab operon, especially the fabD genes [39][42]; and the CRISPRi tool, considered the third promising approach for repressing fab genes and directing carbon flux to malonyl-CoA [43][44]. The positive effect of the ACC1 gene overexpression to increase the malonyl-CoA pool, which was reflected in the increase in resveratrol production, was reported in S. cerevisiae, an E. coli-S. cerevisiae co-culture and Y. lipolytica [84,91,93]. An increasing malonyl-CoA pool was also achieved by the overexpression of a plant malonyl-CoA synthetase (AAE13), resulting in a 2.4-fold increase and accumulation in resveratrol in S. cerevisiae [86].
Increasing the phenylalanine precursor in the prephenic acid pathway was achieved by overexpression of feedback-insensitive alleles encoding DAHP synthase (ScARO4K229L) and chorismate mutase (ScARO7G141S). Applying this strategy combined with the overexpression of the ScACC1S659A, S1157A gene and performing multiple-copies integration of pathways genes (HaTAL, At4CL1, At4CL2, and VvVST1) increased the production to 415.65 and 531.41 mg/L of resveratrol from the final S. cerevisiae strain under fed-batch fermentation with glucose or ethanol as the carbon source, respectively [88]. The same previous strategies were used with the phenylalanine pathway (AtPAL2, AtC4H, At4CL2, and VvVST1) with further metabolic engineering. Overexpression of cytochrome P450 reductase (AtATR2), S. cerevisiae cytochrome B5 (CYB5), and acetyl-CoA synthase (SeACSL641P) was used to increase the precursor supply, as well the deletion of phenylpyruvate decarboxylase (ARO10) to eliminate phenylalanine competing pathways. After this extensive engineering and cultivation in a fed-batch reactor, the strain was able to produce 812 and 755 mg/L of resveratrol from glucose and ethanol, respectively [89].
Y. lipolytica was proven to be a highly promising yeast for producing resveratrol. In Y. lipolytica ST6512, several metabolic strategies were used to finally obtain a strain that produced 12.4 g/L resveratrol, the highest reported resveratrol titer to date from de novo production. In addition to multiple integrations of resveratrol pathway genes (FjTAL, At4CL1, and VvVST1), feedback-insensitive YlARO4K221L and YlARO7G139S were employed to enhance resveratrol production. Further improvement was achieved by optimizing the growth medium and fed-batch fermentation [96].
In bacterial hosts, increased precursor amounts were additionally achieved by overexpression of different genes, such as malonyl-CoA synthetase (matB), malonate carrier protein (matC), tktA, PEP synthetase (ppsA), and acetate assimilation enzyme (acs), or by deleting or inhibiting competing pathway genes such as the pta, ackA, adhE, tyrR, trpR, and fab genes. These strategies might enhance resveratrol production, which requires further experiments on yeast platforms [73,103,104,108,109,110,111].
4. Conclusions
The increased demand for resveratrol for pharmaceutical and cosmetic uses necessitates its production from sustainable sources. Yeast cells are advantageous platforms for resveratrol production because of their fast production, the ease of their cultivation and purification processes, and their remarkable potential for large-scale resveratrol production. Due to their nature as eukaryotic cells and as GRAS organisms, the S. cerevisiae and Y. lipolytica platforms are preferable for resveratrol production. The successful design of a resveratrol production pathway depends on several factors, including determining the appropriate host, examining alternative specific enzymes from various sources, optimizing the codons to maximize the expression of heterologous genes, selecting a convenient transfer gene system (plasmids and their associated genetic elements), and determining favorable culturing parameters. Metabolic engineering has also achieved significant progress in increasing the resveratrol precursors, an important limitation in resveratrol production. Applying all of these elements together in a balanced way as well as testing other strategies that have been applied in E. coli may allow scientists to obtain the desired well-designed resveratrol-producing strain.
This entry is adapted from the peer-reviewed paper 10.3390/biom11060830
References
- Jeandet, P.; Sobarzo-Sánchez, E.; Silva, A.S.; Clément, C.; Nabavi, S.F.; Battino, M.; Rasekhian, M.; Belwal, T.; Habtemariam, S.; Koffas, M. Whole-cell biocatalytic, enzymatic and green chemistry methods for the production of resveratrol and its derivatives. Biotechnol. Adv. 2020, 39, 107461.
- Gugleva, V.; Zasheva, S.; Hristova, M.; Andonova, V. Topical use of resveratrol: Technological aspects. Pharmacia 2020, 67, 89.
- Jeandet, P.; Delaunois, B.; Conreux, A.; Donnez, D.; Nuzzo, V.; Cordelier, S.; Clément, C.; Courot, E. Biosynthesis, metabolism, molecular engineering, and biological functions of stilbene phytoalexins in plants. Biofactors 2010, 36, 331–341.
- Rabesiaka, M.; Rakotondramasy-Rabesiaka, L.; Mabille, I.; Porte, C.; Havet, J.-L. Extraction of trans-resveratrol from red wine and optimization by response surface methodology. Sep. Purif. Technol. 2011, 81, 56–61.
- Jeandet, P.; Douillet-Breuil, A.-C.; Bessis, R.; Debord, S.; Sbaghi, M.; Adrian, M. Phytoalexins from the Vitaceae: Biosynthesis, phytoalexin gene expression in transgenic plants, antifungal activity, and metabolism. J. Agric. Food Chem. 2002, 50, 2731–2741.
- Takaoka, M. Of the phenolic substrate of hellebore (Veratrum grandiflorum Loes. fil.). J. Fac. Sci. Hokkaido Imper. Univ. 1940, 3, 1–16.
- Nonomura, S.; Kanagawa, H.; Makimoto, A. Chemical Constituents of Polygonaceous Plants. I. Studies on the Components of Ko-jo-kon. (Polygonum cuspidatum SIEB. et ZUCC.). Yakugaku Zasshi 1963, 83, 988–990.
- Jeandet, P.; Bessis, R.; Gautheron, B. The production of resveratrol (3, 5, 4’-trihydroxystilbene) by grape berries in different developmental stages. Am. J. Enol. Vitic. 1991, 42, 41–46.
- Lyons, M.M.; Yu, C.; Toma, R.; Cho, S.Y.; Reiboldt, W.; Lee, J.; van Breemen, R. Resveratrol in raw and baked blueberries and bilberries. J. Agric. Food Chem. 2003, 51, 5867–5870.
- Wang, Y.; Catana, F.; Yang, Y.; Roderick, R.; van Breemen, R.B. An LC-MS method for analyzing total resveratrol in grape juice, cranberry juice, and in wine. J. Agric. Food Chem. 2002, 50, 431–435.
- Shrikanta, A.; Kumar, A.; Govindaswamy, V. Resveratrol content and antioxidant properties of underutilized fruits. J. Food Sci. Technol. 2015, 52, 383–390.
- Mudnic, I.; Budimir, D.; Modun, D.; Gunjaca, G.; Generalic, I.; Skroza, D.; Katalinic, V.; Ljubenkov, I.; Boban, M. Antioxidant and vasodilatory effects of blackberry and grape wines. J. Med. Food 2012, 15, 315–321.
- Sales, J.M.; Resurreccion, A.V. Resveratrol in peanuts. Crit. Rev. Food Sci. Nutr. 2014, 54, 734–770.
- Pezzuto, J.M.; Moon, R.C.; Jang, M.-S.; Ouali, A.; Lin, S.; Barillas, K.S. Pharmaceutical Formulations of Resveratrol and Methods of Use Thereof. U.S. Patent US6414037B1, 2 July 2002.
- Ratz-Łyko, A.; Arct, J. Resveratrol as an active ingredient for cosmetic and dermatological applications: A review. J. Cosmet. Laser Ther. 2019, 21, 84–90.
- Nabavi, S.M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316.
- Langcake, P.; Pryce, R.J. The production of resveratrol and the viniferins by grapevines in response to ultraviolet irradiation. Phytochemistry 1977, 16, 1193–1196.
- Jeandet, P.; Delaunois, B.; Aziz, A.; Donnez, D.; Vasserot, Y.; Cordelier, S.; Courot, E. Metabolic engineering of yeast and plants for the production of the biologically active hydroxystilbene, resveratrol. J. Biomed. Biotechnol. 2012, 2012, 1–14.
- Sun, X.; Shen, X.; Jain, R.; Lin, Y.; Wang, J.; Sun, J.; Yan, Y.; Yuan, Q. Synthesis of chemicals by metabolic engineering of microbes. Chem. Soc. Rev. 2015, 44, 3760–3785.
- Braga, A.; Ferreira, P.; Oliveira, J.; Rocha, I.; Faria, N. Heterologous production of resveratrol in bacterial hosts: Current status and perspectives. World J. Microbiol. Biotechnol. 2018, 34, 122.
- Madzak, C. Yarrowia lipolytica: Recent achievements in heterologous protein expression and pathway engineering. Appl. Microbiol. Biotechnol. 2015, 99, 4559–4577.
- Rainha, J.; Gomes, D.; Rodrigues, L.R.; Rodrigues, J.L. Synthetic biology approaches to engineer Saccharomyces cerevisiae towards the industrial production of valuable polyphenolic compounds. Life 2020, 10, 56.
- Thapa, S.B.; Pandey, R.P.; Park, Y.I.; Sohng, J.K. Biotechnological advances in resveratrol production and its chemical diversity. Molecules 2019, 24, 2571.
- Madzak, C. Engineering Yarrowia lipolytica for use in biotechnological applications: A review of major achievements and recent innovations. Mol. Biotechnol. 2018, 60, 621–635.
- Li, M.; Schneider, K.; Kristensen, M.; Borodina, I.; Nielsen, J. Engineering yeast for high-level production of stilbenoid antioxidants. Sci. Rep. 2016, 6, 36827.
- Chen, R.; Yang, S.; Zhang, L.; Zhou, Y.J. Advanced strategies for production of natural products in yeast. iScience 2020, 23, 100879.
- Zhang, Y.; Li, S.-Z.; Li, J.; Pan, X.; Cahoon, R.E.; Jaworski, J.G.; Wang, X.; Jez, J.M.; Chen, F.; Yu, O. Using unnatural protein fusions to engineer resveratrol biosynthesis in yeast and mammalian cells. J. Am. Chem. Soc. 2006, 128, 13030–13031.
- He, Q.; Szczepańska, P.; Yuzbashev, T.; Lazar, Z.; Ledesma-Amaro, R. De novo production of resveratrol from glycerol by engineering different metabolic pathways in Yarrowia lipolytica. Metab. Eng. Commun. 2020, 11, e00146.
- Becker, J.V.; Armstrong, G.O.; Van Der Merwe, M.J.; Lambrechts, M.G.; Vivier, M.A.; Pretorius, I.S. Metabolic engineering of Saccharomyces cerevisiae for the synthesis of the wine-related antioxidant resveratrol. FEMS Yeast Res. 2003, 4, 79–85.
- Wang, Y.; Yu, O. Synthetic scaffolds increased resveratrol biosynthesis in engineered yeast cells. J. Biotechnol. 2012, 157, 258–260.
- Villa-Ruano, N.; Rivera, A.; Rubio-Rosas, E.; Landeta-Cortés, G.; Varela-Caselis, J.L.; Romero-Arenas, O. Comparative activity of six recombinant stilbene synthases in yeast for resveratrol production. Appl. Sci. 2020, 10, 4847.
- Li, M.; Kildegaard, K.R.; Chen, Y.; Rodriguez, A.; Borodina, I.; Nielsen, J. De novo production of resveratrol from glucose or ethanol by engineered Saccharomyces cerevisiae. Metab. Eng. 2015, 32, 1–11.
- Yuan, S.-F.; Yi, X.; Johnston, T.G.; Alper, H.S. De novo resveratrol production through modular engineering of an Escherichia coli–Saccharomyces cerevisiae co-culture. Microb. Cell Factories 2020, 19, 143.
- Palmer, C.M.; Miller, K.K.; Nguyen, A.; Alper, H.S. Engineering 4-coumaroyl-CoA derived polyketide production in Yarrowia lipolytica through a β-oxidation mediated strategy. Metab. Eng. 2020, 57, 174–181.
- Gu, Y.; Ma, J.; Zhu, Y.; Ding, X.; Xu, P. Engineering Yarrowia lipolytica as a chassis for de novo synthesis of five aromatic-derived natural products and chemicals. ACS Synth. Biol. 2020, 9, 2096–2106.
- Sáez-Sáez, J.; Wang, G.; Marella, E.R.; Sudarsan, S.; Cernuda Pastor, M.; Borodina, I. Engineering the oleaginous yeast Yarrowia lipolytica for high-level resveratrol production. Metab. Eng. 2020, 62, 51–61.
- Sydor, T.; Schaffer, S.; Boles, E. Considerable increase in resveratrol production by recombinant industrial yeast strains with use of rich medium. Appl. Environ. Microbiol. 2010, 76, 3361–3363.
- Shrestha, A.; Pandey, R.P.; Sohng, J.K. Biosynthesis of resveratrol and piceatannol in engineered microbial strains: Achievements and perspectives. Appl. Microbiol. Biotechnol. 2019, 103, 2959–2972.
- Yang, Y.; Lin, Y.; Li, L.; Linhardt, R.J.; Yan, Y. Regulating malonyl-CoA metabolism via synthetic antisense RNAs for enhanced biosynthesis of natural products. Metab. Eng. 2015, 29, 217–226.
- Lu, Y.; Shao, D.; Shi, J.; Huang, Q.; Yang, H.; Jin, M. Strategies for enhancing resveratrol production and the expression of pathway enzymes. Appl. Microbiol. Biotechnol. 2016, 100, 7407–7421.
- Lim, C.G.; Fowler, Z.L.; Hueller, T.; Schaffer, S.; Koffas, M.A. High-yield resveratrol production in engineered Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 3451–3460.
- Wu, J.; Yu, O.; Du, G.; Zhou, J.; Chen, J. Fine-tuning of the fatty acid pathway by synthetic antisense RNA for enhanced (2S)-naringenin production from L-tyrosine in Escherichia coli. Appl. Environ. Microbiol. 2014, 80, 7283–7292.
- Liang, J.-l.; Guo, L.-q.; Lin, J.-f.; He, Z.-q.; Cai, F.-j.; Chen, J.-f. A novel process for obtaining pinosylvin using combinatorial bioengineering in Escherichia coli. World J. Microbiol. Biotechnol. 2016, 32, 102.
- Wu, J.; Du, G.; Chen, J.; Zhou, J. Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Sci. Rep. 2015, 5, 1–14.
