In human cancer, circular RNAs (circRNAs) were implicated in the control of oncogenic activities such as tumor cell proliferation, epithelial-mesenchymal transition, invasion, metastasis and chemoresistance. The most widely described mechanism of action of circRNAs is their ability to act as competing endogenous RNAs (ceRNAs) for miRNAs, lncRNAs and mRNAs, thus impacting along their axis, despite the fact that a variety of additional mechanisms of action are emerging, representing an open and expanding field of study.
- circRNA
- cancer
- backsplicing
- host gene
- parental gene
- translation of ncRNA
- miRNA
1. Overview
Circular RNAs (circRNAs) belong to a new class of non-coding RNAs implicated in cellular physiological functions but also in the evolution of various human pathologies. Due to their circular shape, circRNAs are resistant to degradation by exonuclease activity, making them more stable than linear RNAs. Several findings reported that circRNAs are aberrantly modulated in human cancer tissues, thus affecting carcinogenesis and metastatization. We aim to report the most recent and relevant results about novel circRNA functions and molecular regulation, to dissert about their role as reliable cancer biomarkers, and to hypothesize their contribution to multiple hallmarks of cancer.
Next generation RNA sequencing techniques, implemented in the recent years, have allowed us to identify circular RNAs (circRNAs), covalently closed loop structures resulting in RNA molecules that are more stable than linear RNAs. This class of non-coding RNA is emerging to be involved in a variety of cell functions during development, differentiation, and in many diseases, including cancer. Among the described biological activities, circRNAs have been implicated in microRNA (miRNA) sequestration, modulation of protein–protein interactions and regulation of mRNA transcription. In human cancer, circRNAs were implicated in the control of oncogenic activities such as tumor cell proliferation, epithelial-mesenchymal transition, invasion, metastasis and chemoresistance. The most widely described mechanism of action of circRNAs is their ability to act as competing endogenous RNAs (ceRNAs) for miRNAs, lncRNAs and mRNAs, thus impacting along their axis, despite the fact that a variety of additional mechanisms of action are emerging, representing an open and expanding field of study. Furthermore, research is currently focusing on understanding the possible implications of circRNAs in diagnostics, prognosis prediction, effectiveness of therapies and, eventually, therapeutic intervention in human cancer. The purpose of this review is to discuss new knowledge on the mechanisms of circRNA action, beyond ceRNA, their impact on human cancer and to dissect their potential value as biomarkers and therapeutic targets.
2. circRNAs
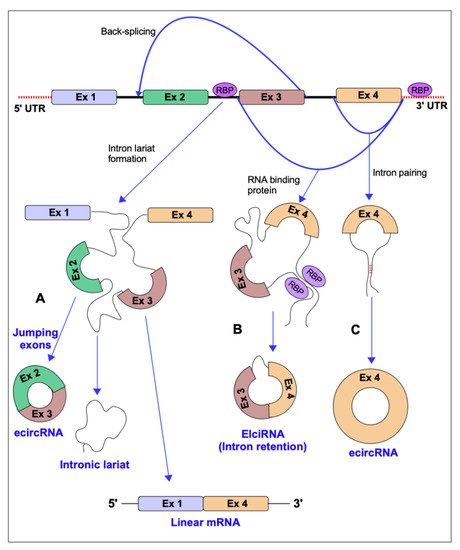
3. Conclusions
This entry is adapted from the peer-reviewed paper 10.3390/cancers13133154
References
- Volovat, S.R.; Volovat, C.; Hordila, I.; Hordila, D.A.; Mirestean, C.C.; Miron, O.T.; Lungulescu, C.; Scripcariu, D.V.; Stolniceanu, C.R.; Konsoulova-Kirova, A.A.; et al. MiRNA and LncRNA as Potential Biomarkers in Triple-Negative Breast Cancer: A Review. Front. Oncol. 2020, 10, 526850.
- Zhang, Z.; Zhang, J.; Diao, L.; Han, L. Small non-coding RNAs in human cancer: Function, clinical utility, and characterization. Oncogene 2021, 40, 1570–1577.
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856.
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338.
- Nisar, S.; Bhat, A.A.; Singh, M.; Karedath, T.; Rizwan, A.; Hashem, S.; Bagga, P.; Reddy, R.; Jamal, F.; Uddin, S.; et al. Insights Into the Role of CircRNAs: Biogenesis, Characterization, Functional, and Clinical Impact in Human Malignancies. Front. Cell Dev. Biol. 2021, 9, 617281.
- Szabo, L.; Salzman, J. Detecting circular RNAs: Bioinformatic and experimental challenges. Nat. Rev. Genet. 2016, 17, 679–692.
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; Robinson, D.R.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e813.
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015, 10, 170–177.
- Tang, X.; Ren, H.; Guo, M.; Qian, J.; Yang, Y.; Gu, C. Review on circular RNAs and new insights into their roles in cancer. Comput. Struct. Biotechnol. J. 2021, 19, 910–928.
- Wang, P.L.; Bao, Y.; Yee, M.C.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Salzman, J. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 2014, 9, e90859.
- Zhang, P.; Li, S.; Chen, M. Characterization and Function of Circular RNAs in Plants. Front. Mol. Biosci. 2020, 7, 91.
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147.
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66.
- Eger, N.; Schoppe, L.; Schuster, S.; Laufs, U.; Boeckel, J.N. Circular RNA Splicing. Adv. Exp. Med. Biol. 2018, 1087, 41–52.
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264.
- Kristensen, L.S.; Hansen, T.B.; Veno, M.T.; Kjems, J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 2018, 37, 555–565.
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461.
- Li, Y.; Feng, W.; Kong, M.; Liu, R.; Wu, A.; Shen, L.; Tang, Z.; Wang, F. Exosomal circRNAs: A new star in cancer. Life Sci. 2021, 269, 119039.
- Li, J.; Sun, D.; Pu, W.; Wang, J.; Peng, Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer 2020, 6, 319–336.
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691.
- Dumache, R. Early Diagnosis of Oral Squamous Cell Carcinoma by Salivary microRNAs. Clin. Lab. 2017, 63, 1771–1776.
- Liu, Y.; He, A.; Liu, B.; Huang, Z.; Mei, H. Potential Role of lncRNA H19 as a Cancer Biomarker in Human Cancers Detection and Diagnosis: A Pooled Analysis Based on 1585 Subjects. BioMed Res. Int. 2019, 2019, 9056458.
- Luzon-Toro, B.; Fernandez, R.M.; Martos-Martinez, J.M.; Rubio-Manzanares-Dorado, M.; Antinolo, G.; Borrego, S. LncRNA LUCAT1 as a novel prognostic biomarker for patients with papillary thyroid cancer. Sci. Rep. 2019, 9, 14374.
