In human cancer, circular RNAs (circRNAs) were implicated in the control of oncogenic activities such as tumor cell proliferation, epithelial-mesenchymal transition, invasion, metastasis and chemoresistance. The most widely described mechanism of action of circRNAs is their ability to act as competing endogenous RNAs (ceRNAs) for miRNAs, lncRNAs and mRNAs, thus impacting along their axis, despite the fact that a variety of additional mechanisms of action are emerging, representing an open and expanding field of study.
- circRNA
- cancer
- backsplicing
- host gene
- parental gene
- translation of ncRNA
- miRNA
1. Overview
Circular RNAs (circRNAs) belong to a new class of non-coding RNAs implicated in cellular physiological functions but also in the evolution of various human pathologies. Due to their circular shape, circRNAs are resistant to degradation by exonuclease activity, making them more stable than linear RNAs. Several findings reported that circRNAs are aberrantly modulated in human cancer tissues, thus affecting carcinogenesis and metastatization. We aim to report the most recent and relevant results about novel circRNA functions and molecular regulation, to dissert about their role as reliable cancer biomarkers, and to hypothesize their contribution to multiple hallmarks of cancer.
Next generation RNA sequencing techniques, implemented in the recent years, have allowed us to identify circular RNAs (circRNAs), covalently closed loop structures resulting in RNA molecules that are more stable than linear RNAs. This class of non-coding RNA is emerging to be involved in a variety of cell functions during development, differentiation, and in many diseases, including cancer. Among the described biological activities, circRNAs have been implicated in microRNA (miRNA) sequestration, modulation of protein–protein interactions and regulation of mRNA transcription. In human cancer, circRNAs were implicated in the control of oncogenic activities such as tumor cell proliferation, epithelial-mesenchymal transition, invasion, metastasis and chemoresistance. The most widely described mechanism of action of circRNAs is their ability to act as competing endogenous RNAs (ceRNAs) for miRNAs, lncRNAs and mRNAs, thus impacting along their axis, despite the fact that a variety of additional mechanisms of action are emerging, representing an open and expanding field of study. Furthermore, research is currently focusing on understanding the possible implications of circRNAs in diagnostics, prognosis prediction, effectiveness of therapies and, eventually, therapeutic intervention in human cancer. The purpose of this review is to discuss new knowledge on the mechanisms of circRNA action, beyond ceRNA, their impact on human cancer and to dissect their potential value as biomarkers and therapeutic targets.
2. circRNAs
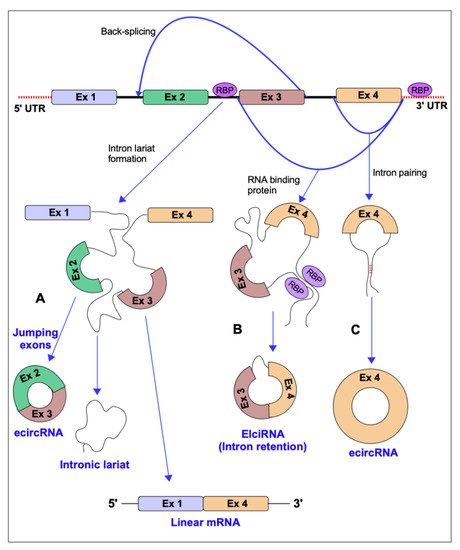
3. Conclusions
Volovat, S.R.; Volovat, C.; Hordila, I.; Hordila, D.A.; Mirestean, C.C.; Miron, O.T.; Lungulescu, C.; Scripcariu, D.V.; Stolniceanu, C.R.; Konsoulova-Kirova, A.A.; et al. MiRNA and LncRNA as Potential Biomarkers in Triple-Negative Breast Cancer: A Review. Frontiers in oncology 2020, 10, 526850.
- Zhang, Z.; Zhang, J.; Diao, L.; Han, L. Small non-coding RNAs in human cancer: function, clinical utility, and characterization. Oncogene 2021, 40, 1570-1577.
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proceedings of the National Academy of Sciences of the United States of America 1976, 73, 3852-3856.
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333-338.
- Nisar, S.; Bhat, A.A.; Singh, M.; Karedath, T.; Rizwan, A.; Hashem, S.; Bagga, P.; Reddy, R.; Jamal, F.; Uddin, S.; et al. Insights Into the Role of CircRNAs: Biogenesis, Characterization, Functional, and Clinical Impact in Human Malignancies. Frontiers in cell and developmental biology 2021, 9, 617281.
- Szabo, L.; Salzman, J. Detecting circular RNAs: bioinformatic and experimental challenges. Nature reviews. Genetics 2016, 17, 679-692.
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; Robinson, D.R.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869-881 e813.
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell reports 2015, 10, 170-177.
- Tang, X.; Ren, H.; Guo, M.; Qian, J.; Yang, Y.; Gu, C. Review on circular RNAs and new insights into their roles in cancer. Computational and structural biotechnology journal 2021, 19, 910-928.
- Wang, P.L.; Bao, Y.; Yee, M.C.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Salzman, J. Circular RNA is expressed across the eukaryotic tree of life. PloS one 2014, 9, e90859.
- Zhang, P.; Li, S.; Chen, M. Characterization and Function of Circular RNAs in Plants. Frontiers in molecular biosciences 2020, 7, 91.
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134-147.
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Molecular cell 2014, 56, 55-66.
- Eger, N.; Schoppe, L.; Schuster, S.; Laufs, U.; Boeckel, J.N. Circular RNA Splicing. Advances in experimental medicine and biology 2018, 1087, 41-52.
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nature structural & molecular biology 2015, 22, 256-264.
- Kristensen, L.S.; Hansen, T.B.; Veno, M.T.; Kjems, J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 2018, 37, 555-565.
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nature biotechnology 2014, 32, 453-461.
- Li, Y.; Feng, W.; Kong, M.; Liu, R.; Wu, A.; Shen, L.; Tang, Z.; Wang, F. Exosomal circRNAs: A new star in cancer. Life sciences 2021, 269, 119039.
- Wawrzyniak, O.; Zarebska, Z.; Kuczynski, K.; Gotz-Wieckowska, A.; Rolle, K. Protein-Related Circular RNAs in Human Pathologies. Cells 2020, 9.
- Lai, D.; Proctor, J.R.; Meyer, I.M. On the importance of cotranscriptional RNA structure formation. RNA 2013, 19, 1461-1473.
- Sengupta, A.; Sung, H.L.; Nesbitt, D.J. Amino Acid Specific Effects on RNA Tertiary Interactions: Single-Molecule Kinetic and Thermodynamic Studies. The journal of physical chemistry. B 2016, 120, 10615-10627.
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell death and differentiation 2017, 24, 357-370.
- Yang, Q.; Du, W.W.; Wu, N.; Yang, W.; Awan, F.M.; Fang, L.; Ma, J.; Li, X.; Zeng, Y.; Yang, Z.; et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell death and differentiation 2017, 24, 1609-1620.
- Yang, Z.G.; Awan, F.M.; Du, W.W.; Zeng, Y.; Lyu, J.; Wu; Gupta, S.; Yang, W.; Yang, B.B. The Circular RNA Interacts with STAT3, Increasing Its Nuclear Translocation and Wound Repair by Modulating Dnmt3a and miR-17 Function. Molecular therapy : the journal of the American Society of Gene Therapy 2017, 25, 2062-2074.
- Du, W.W.; Yang, W.; Chen, Y.; Wu, Z.K.; Foster, F.S.; Yang, Z.; Li, X.; Yang, B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. European heart journal 2017, 38, 1402-1412.
- Abdelmohsen, K.; Panda, A.C.; Munk, R.; Grammatikakis, I.; Dudekula, D.B.; De, S.; Kim, J.; Noh, J.H.; Kim, K.M.; Martindale, J.L.; et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA biology 2017, 14, 361-369.
- Tsitsipatis, D.; Grammatikakis, I.; Driscoll, R.K.; Yang, X.; Abdelmohsen, K.; Harris, S.C.; Yang, J.H.; Herman, A.B.; Chang, M.W.; Munk, R.; et al. AUF1 ligand circPCNX reduces cell proliferation by competing with p21 mRNA to increase p21 production. Nucleic acids research 2021, 49, 1631-1646.
- Chen, L.; Kong, R.; Wu, C.; Wang, S.; Liu, Z.; Liu, S.; Li, S.; Chen, T.; Mao, C. Circ-MALAT1 Functions as Both an mRNA Translation Brake and a microRNA Sponge to Promote Self-Renewal of Hepatocellular Cancer Stem Cells. Adv Sci (Weinh) 2020, 7, 1900949.
- Chen, R.X.; Chen, X.; Xia, L.P.; Zhang, J.X.; Pan, Z.Z.; Ma, X.D.; Han, K.; Chen, J.W.; Judde, J.G.; Deas, O.; et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nature communications 2019, 10, 4695.
- Wu, N.; Yuan, Z.; Du, K.Y.; Fang, L.; Lyu, J.; Zhang, C.; He, A.; Eshaghi, E.; Zeng, K.; Ma, J.; et al. Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell death and differentiation 2019, 26, 2758-2773.
- Guarnerio, J.; Zhang, Y.; Cheloni, G.; Panella, R.; Mae Katon, J.; Simpson, M.; Matsumoto, A.; Papa, A.; Loretelli, C.; Petri, A.; et al. Intragenic antagonistic roles of protein and circRNA in tumorigenesis. Cell research 2019, 29, 628-640.
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: metabolism, functions and interactions with proteins. Molecular cancer 2020, 19, 172.
- Myatt, S.S.; Lam, E.W. The emerging roles of forkhead box (Fox) proteins in cancer. Nature reviews. Cancer 2007, 7, 847-859.
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic acids research 2016, 44, 2846-2858.
- Fang, L.; Du, W.W.; Lyu, J.; Dong, J.; Zhang, C.; Yang, W.; He, A.; Kwok, Y.S.S.; Ma, J.; Wu, N.; et al. Enhanced breast cancer progression by mutant p53 is inhibited by the circular RNA circ-Ccnb1. Cell death and differentiation 2018, 25, 2195-2208.
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nature communications 2016, 7, 12429.
- Hu, X.; Wu, D.; He, X.; Zhao, H.; He, Z.; Lin, J.; Wang, K.; Wang, W.; Pan, Z.; Lin, H.; et al. circGSK3beta promotes metastasis in esophageal squamous cell carcinoma by augmenting beta-catenin signaling. Molecular cancer 2019, 18, 160.
- Ding, L.; Zhao, Y.; Dang, S.; Wang, Y.; Li, X.; Yu, X.; Li, Z.; Wei, J.; Liu, M.; Li, G. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Molecular cancer 2019, 18, 45.
- Wang, L.; Long, H.; Zheng, Q.; Bo, X.; Xiao, X.; Li, B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Molecular cancer 2019, 18, 119.
- Kwek, K.Y.; Murphy, S.; Furger, A.; Thomas, B.; O'Gorman, W.; Kimura, H.; Proudfoot, N.J.; Akoulitchev, A. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nature structural biology 2002, 9, 800-805.
- Feng, Y.; Yang, Y.; Zhao, X.; Fan, Y.; Zhou, L.; Rong, J.; Yu, Y. Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell death & disease 2019, 10, 792.
- Chen, N.; Zhao, G.; Yan, X.; Lv, Z.; Yin, H.; Zhang, S.; Song, W.; Li, X.; Li, L.; Du, Z.; et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome biology 2018, 19, 218.
- Li, H.; Yang, F.; Hu, A.; Wang, X.; Fang, E.; Chen, Y.; Li, D.; Song, H.; Wang, J.; Guo, Y.; et al. Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO molecular medicine 2019, 11, e10835.
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Molecular cell 2013, 51, 792-806.
- Payne, J.M.; Laybourn, P.J.; Dahmus, M.E. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. The Journal of biological chemistry 1989, 264, 19621-19629.
- Conlon, E.G.; Manley, J.L. RNA-binding proteins in neurodegeneration: mechanisms in aggregate. Genes & development 2017, 31, 1509-1528.
- Zhao, W.; Wang, S.; Qin, T.; Wang, W. Circular RNA (circ-0075804) promotes the proliferation of retinoblastoma via combining heterogeneous nuclear ribonucleoprotein K (HNRNPK) to improve the stability of E2F transcription factor 3 E2F3. Journal of cellular biochemistry 2020, 121, 3516-3525.
- Liu, B.; Yang, G.; Wang, X.; Liu, J.; Lu, Z.; Wang, Q.; Xu, B.; Liu, Z.; Li, J. CircBACH1 (hsa_circ_0061395) promotes hepatocellular carcinoma growth by regulating p27 repression via HuR. Journal of cellular physiology 2020, 235, 6929-6941.
- Lei, M.; Zheng, G.; Ning, Q.; Zheng, J.; Dong, D. Translation and functional roles of circular RNAs in human cancer. Molecular cancer 2020, 19, 30.
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Molecular cell 2017, 66, 22-37 e29.
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Chen, L.L.; Wang, Y.; Wong, C.C.; et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell research 2017, 27, 626-641.
- Li, H.; Li, Q.; He, S. Hsa_circ_0025202 suppresses cell tumorigenesis and tamoxifen resistance via miR-197-3p/HIPK3 axis in breast cancer. World journal of surgical oncology 2021, 19, 39.
- Liang, W.C.; Wong, C.W.; Liang, P.P.; Shi, M.; Cao, Y.; Rao, S.T.; Tsui, S.K.; Waye, M.M.; Zhang, Q.; Fu, W.M.; et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome biology 2019, 20, 84.
- Yang, Y.; Gao, X.; Zhang, M.; Yan, S.; Sun, C.; Xiao, F.; Huang, N.; Yang, X.; Zhao, K.; Zhou, H.; et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. Journal of the National Cancer Institute 2018, 110.
- Ye, F.; Gao, G.; Zou, Y.; Zheng, S.; Zhang, L.; Ou, X.; Xie, X.; Tang, H. circFBXW7 Inhibits Malignant Progression by Sponging miR-197-3p and Encoding a 185-aa Protein in Triple-Negative Breast Cancer. Molecular therapy. Nucleic acids 2019, 18, 88-98.
- Wesselhoeft, R.A.; Kowalski, P.S.; Parker-Hale, F.C.; Huang, Y.; Bisaria, N.; Anderson, D.G. RNA Circularization Diminishes Immunogenicity and Can Extend Translation Duration In Vivo. Molecular cell 2019, 74, 508-520 e504.
- Chen, Y.G.; Chen, R.; Ahmad, S.; Verma, R.; Kasturi, S.P.; Amaya, L.; Broughton, J.P.; Kim, J.; Cadena, C.; Pulendran, B.; et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Molecular cell 2019, 76, 96-109 e109.
- Zhang, X.L.; Xu, L.L.; Wang, F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell biology international 2017, 41, 1056-1064.
- Wei, C.Y.; Zhu, M.X.; Lu, N.H.; Liu, J.Q.; Yang, Y.W.; Zhang, Y.; Shi, Y.D.; Feng, Z.H.; Li, J.X.; Qi, F.Z.; et al. Circular RNA circ_0020710 drives tumor progression and immune evasion by regulating the miR-370-3p/CXCL12 axis in melanoma. Molecular cancer 2020, 19, 84.
- Huang, X.Y.; Zhang, P.F.; Wei, C.Y.; Peng, R.; Lu, J.C.; Gao, C.; Cai, J.B.; Yang, X.; Fan, J.; Ke, A.W.; et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Molecular cancer 2020, 19, 92.
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O'Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America 2008, 105, 10513-10518.
- Pant, S.; Hilton, H.; Burczynski, M.E. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochemical pharmacology 2012, 83, 1484-1494.
- Kalluri, R. The biology and function of exosomes in cancer. The Journal of clinical investigation 2016, 126, 1208-1215.
- Maia, J.; Caja, S.; Strano Moraes, M.C.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Frontiers in cell and developmental biology 2018, 6, 18.
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Developmental cell 2019, 49, 347-360.
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell research 2015, 25, 981-984.
- Dou, Y.; Cha, D.J.; Franklin, J.L.; Higginbotham, J.N.; Jeppesen, D.K.; Weaver, A.M.; Prasad, N.; Levy, S.; Coffey, R.J.; Patton, J.G.; et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Scientific reports 2016, 6, 37982.
- Wang, M.; Yu, F.; Li, P.; Wang, K. Emerging Function and Clinical Significance of Exosomal circRNAs in Cancer. Molecular therapy. Nucleic acids 2020, 21, 367-383.
- Di Agostino, S.; Riccioli, A.; De Cesaris, P.; Fontemaggi, G.; Blandino, G.; Filippini, A.; Fazi, F. Circular RNAs in Embryogenesis and Cell Differentiation With a Focus on Cancer Development. Frontiers in cell and developmental biology 2020, 8, 389.
- Hansen, T.B.; Kjems, J.; Damgaard, C.K. Circular RNA and miR-7 in cancer. Cancer research 2013, 73, 5609-5612.
- Li, Z.; Yanfang, W.; Li, J.; Jiang, P.; Peng, T.; Chen, K.; Zhao, X.; Zhang, Y.; Zhen, P.; Zhu, J.; et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer letters 2018, 432, 237-250.
- Limb, C.; Liu, D.S.K.; Veno, M.T.; Rees, E.; Krell, J.; Bagwan, I.N.; Giovannetti, E.; Pandha, H.; Strobel, O.; Rockall, T.A.; et al. The Role of Circular RNAs in Pancreatic Ductal Adenocarcinoma and Biliary-Tract Cancers. Cancers 2020, 12.
- Li, J.; Li, Z.; Jiang, P.; Peng, M.; Zhang, X.; Chen, K.; Liu, H.; Bi, H.; Liu, X.; Li, X. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. Journal of experimental & clinical cancer research : CR 2018, 37, 177.
- Wang, S.; Su, W.; Zhong, C.; Yang, T.; Chen, W.; Chen, G.; Liu, Z.; Wu, K.; Zhong, W.; Li, B.; et al. An Eight-CircRNA Assessment Model for Predicting Biochemical Recurrence in Prostate Cancer. Frontiers in cell and developmental biology 2020, 8, 599494.
- Dall'Era, M.A.; Albertsen, P.C.; Bangma, C.; Carroll, P.R.; Carter, H.B.; Cooperberg, M.R.; Freedland, S.J.; Klotz, L.H.; Parker, C.; Soloway, M.S. Active surveillance for prostate cancer: a systematic review of the literature. European urology 2012, 62, 976-983.
- Zhang, N.; Nan, A.; Chen, L.; Li, X.; Jia, Y.; Qiu, M.; Dai, X.; Zhou, H.; Zhu, J.; Zhang, H.; et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Molecular cancer 2020, 19, 101.
- Mao, Y.; Yang, D.; He, J.; Krasna, M.J. Epidemiology of Lung Cancer. Surgical oncology clinics of North America 2016, 25, 439-445.
- Lin, S.; Xiong, W.; Liu, H.; Pei, L.; Yi, H.; Guan, Y. Profiling and Integrated Analysis of Differentially Expressed Circular RNAs in Plasma Exosomes as Novel Biomarkers for Advanced-Stage Lung Adenocarcinoma. OncoTargets and therapy 2020, 13, 12965-12977.
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA: a cancer journal for clinicians 2018, 68, 284-296.
- Motohara, T.; Masuda, K.; Morotti, M.; Zheng, Y.; El-Sahhar, S.; Chong, K.Y.; Wietek, N.; Alsaadi, A.; Karaminejadranjbar, M.; Hu, Z.; et al. An evolving story of the metastatic voyage of ovarian cancer cells: cellular and molecular orchestration of the adipose-rich metastatic microenvironment. Oncogene 2019, 38, 2885-2898.
- Zong, Z.H.; Du, Y.P.; Guan, X.; Chen, S.; Zhao, Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. Journal of experimental & clinical cancer research : CR 2019, 38, 437.
- Wu, H.; Xiao, Z.; Wang, K.; Liu, W.; Hao, Q. MiR-145 is downregulated in human ovarian cancer and modulates cell growth and invasion by targeting p70S6K1 and MUC1. Biochemical and biophysical research communications 2013, 441, 693-700.
- Lyu, P.; Zhai, Z.; Hao, Z.; Zhang, H.; He, J. CircWHSC1 serves as an oncogene to promote hepatocellular carcinoma progression. European journal of clinical investigation 2021, e13487.
- Hu, W.; Liu, C.; Bi, Z.Y.; Zhou, Q.; Zhang, H.; Li, L.L.; Zhang, J.; Zhu, W.; Song, Y.Y.; Zhang, F.; et al. Comprehensive landscape of extracellular vesicle-derived RNAs in cancer initiation, progression, metastasis and cancer immunology. Molecular cancer 2020, 19, 102.
- Liu, Y.; Yang, Y.; Wang, Z.; Fu, X.; Chu, X.M.; Li, Y.; Wang, Q.; He, X.; Li, M.; Wang, K.; et al. Insights into the regulatory role of circRNA in angiogenesis and clinical implications. Atherosclerosis 2020, 298, 14-26.
- Ou, Z.L.; Luo, Z.; Wei, W.; Liang, S.; Gao, T.L.; Lu, Y.B. Hypoxia-induced shedding of MICA and HIF1A-mediated immune escape of pancreatic cancer cells from NK cells: role of circ_0000977/miR-153 axis. RNA biology 2019, 16, 1592-1603.
- Yarla, N.S.; Madka, V.; Pathuri, G.; Rao, C.V. Molecular Targets in Precision Chemoprevention of Colorectal Cancer: An Update from Pre-Clinical to Clinical Trials. International journal of molecular sciences 2020, 21.
- Hon, K.W.; Ab-Mutalib, N.S.; Abdullah, N.M.A.; Jamal, R.; Abu, N. Extracellular Vesicle-derived circular RNAs confers chemoresistance in Colorectal cancer. Scientific reports 2019, 9, 16497.
- Sun, Y.; Cao, Z.; Shan, J.; Gao, Y.; Liu, X.; Ma, D.; Li, Z. Hsa_circ_0020095 Promotes Oncogenesis and Cisplatin Resistance in Colon Cancer by Sponging miR-487a-3p and Modulating SOX9. Frontiers in cell and developmental biology 2020, 8, 604869.
- Xu, Y.; Qiu, A.; Peng, F.; Tan, X.; Wang, J.; Gong, X. Exosomal transfer of circular RNA FBXW7 ameliorates the chemoresistance to oxaliplatin in colorectal cancer by sponging miR-18b-5p. Neoplasma 2021, 68, 108-118.
- Wang, X.; Zhang, H.; Yang, H.; Bai, M.; Ning, T.; Deng, T.; Liu, R.; Fan, Q.; Zhu, K.; Li, J.; et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Molecular oncology 2020, 14, 539-555.
- Zheng, S.R.; Huang, Q.D.; Zheng, Z.H.; Zhang, Z.T.; Guo, G.L. circGFRA1 affects the sensitivity of triple negative breast cancer cells to paclitaxel via the miR-361-5p/TLR4 pathway. Journal of biochemistry 2021.
- Li, M.; Cai, J.; Han, X.; Ren, Y. Downregulation of circNRIP1 Suppresses the Paclitaxel Resistance of Ovarian Cancer via Regulating the miR-211-5p/HOXC8 Axis. Cancer management and research 2020, 12, 9159-9171.
- Luo, Y.; Gui, R. Circulating exosomal circFoxp1 confers cisplatin resistance in epithelial ovarian cancer cells. Journal of gynecologic oncology 2020, 31, e75.
- Teow, H.M.; Zhou, Z.; Najlah, M.; Yusof, S.R.; Abbott, N.J.; D'Emanuele, A. Delivery of paclitaxel across cellular barriers using a dendrimer-based nanocarrier. International journal of pharmaceutics 2013, 441, 701-711.
- Xu, J.; Ni, L.; Zhao, F.; Dai, X.; Tao, J.; Pan, J.; Shi, A.; Shen, Z.; Su, C.; Zhang, Y. Overexpression of hsa_circ_0002874 promotes resistance of non-small cell lung cancer to paclitaxel by modulating miR-1273f/MDM2/p53 pathway. Aging 2021, 13, 5986-6009.
- Xu, X.; Tao, R.; Sun, L.; Ji, X. Exosome-transferred hsa_circ_0014235 promotes DDP chemoresistance and deteriorates the development of non-small cell lung cancer by mediating the miR-520a-5p/CDK4 pathway. Cancer cell international 2020, 20, 552.
- Huang, M.S.; Liu, J.Y.; Xia, X.B.; Liu, Y.Z.; Li, X.; Yin, J.Y.; Peng, J.B.; Wu, L.; Zhang, W.; Zhou, H.H.; et al. Hsa_circ_0001946 Inhibits Lung Cancer Progression and Mediates Cisplatin Sensitivity in Non-small Cell Lung Cancer via the Nucleotide Excision Repair Signaling Pathway. Frontiers in oncology 2019, 9, 508.
- Siegel, R.; Desantis, C.; Jemal, A. Colorectal cancer statistics, 2014. CA: a cancer journal for clinicians 2014, 64, 104-117.
- Llovet, J.M.; Montal, R.; Sia, D.; Finn, R.S. Molecular therapies and precision medicine for hepatocellular carcinoma. Nature reviews. Clinical oncology 2018, 15, 599-616.
- Weng, H.; Zeng, L.; Cao, L.; Chen, T.; Li, Y.; Xu, Y.; Peng, Y.; Ye, Y. circFOXM1 contributes to sorafenib resistance of hepatocellular carcinoma cells by regulating MECP2 via miR-1324. Molecular therapy. Nucleic acids 2021, 23, 811-820.
- Mantovani, S.; Oliviero, B.; Varchetta, S.; Mele, D.; Mondelli, M.U. Natural Killer Cell Responses in Hepatocellular Carcinoma: Implications for Novel Immunotherapeutic Approaches. Cancers 2020, 12.
- Zhang, P.F.; Gao, C.; Huang, X.Y.; Lu, J.C.; Guo, X.J.; Shi, G.M.; Cai, J.B.; Ke, A.W. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Molecular cancer 2020, 19, 110.
- Li, J.; Sun, D.; Pu, W.; Wang, J.; Peng, Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends in cancer 2020, 6, 319-336.
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nature reviews. Genetics 2019, 20, 675-691.
- Dumache, R. Early Diagnosis of Oral Squamous Cell Carcinoma by Salivary microRNAs. Clinical laboratory 2017, 63, 1771-1776.
- Liu, Y.; He, A.; Liu, B.; Huang, Z.; Mei, H. Potential Role of lncRNA H19 as a Cancer Biomarker in Human Cancers Detection and Diagnosis: A Pooled Analysis Based on 1585 Subjects. BioMed research international 2019, 2019, 9056458.
- Luzon-Toro, B.; Fernandez, R.M.; Martos-Martinez, J.M.; Rubio-Manzanares-Dorado, M.; Antinolo, G.; Borrego, S. LncRNA LUCAT1 as a novel prognostic biomarker for patients with papillary thyroid cancer. Scientific reports 2019, 9, 14374.
This entry is adapted from the peer-reviewed paper 10.3390/cancers13133154
References
- Volovat, S.R.; Volovat, C.; Hordila, I.; Hordila, D.A.; Mirestean, C.C.; Miron, O.T.; Lungulescu, C.; Scripcariu, D.V.; Stolniceanu, C.R.; Konsoulova-Kirova, A.A.; et al. MiRNA and LncRNA as Potential Biomarkers in Triple-Negative Breast Cancer: A Review. Front. Oncol. 2020, 10, 526850.
- Zhang, Z.; Zhang, J.; Diao, L.; Han, L. Small non-coding RNAs in human cancer: Function, clinical utility, and characterization. Oncogene 2021, 40, 1570–1577.
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856.
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338.
- Nisar, S.; Bhat, A.A.; Singh, M.; Karedath, T.; Rizwan, A.; Hashem, S.; Bagga, P.; Reddy, R.; Jamal, F.; Uddin, S.; et al. Insights Into the Role of CircRNAs: Biogenesis, Characterization, Functional, and Clinical Impact in Human Malignancies. Front. Cell Dev. Biol. 2021, 9, 617281.
- Szabo, L.; Salzman, J. Detecting circular RNAs: Bioinformatic and experimental challenges. Nat. Rev. Genet. 2016, 17, 679–692.
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; Robinson, D.R.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e813.
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015, 10, 170–177.
- Tang, X.; Ren, H.; Guo, M.; Qian, J.; Yang, Y.; Gu, C. Review on circular RNAs and new insights into their roles in cancer. Comput. Struct. Biotechnol. J. 2021, 19, 910–928.
- Wang, P.L.; Bao, Y.; Yee, M.C.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Salzman, J. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 2014, 9, e90859.
- Zhang, P.; Li, S.; Chen, M. Characterization and Function of Circular RNAs in Plants. Front. Mol. Biosci. 2020, 7, 91.
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147.
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66.
- Eger, N.; Schoppe, L.; Schuster, S.; Laufs, U.; Boeckel, J.N. Circular RNA Splicing. Adv. Exp. Med. Biol. 2018, 1087, 41–52.
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264.
- Kristensen, L.S.; Hansen, T.B.; Veno, M.T.; Kjems, J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 2018, 37, 555–565.
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461.
- Li, Y.; Feng, W.; Kong, M.; Liu, R.; Wu, A.; Shen, L.; Tang, Z.; Wang, F. Exosomal circRNAs: A new star in cancer. Life Sci. 2021, 269, 119039.
- Li, J.; Sun, D.; Pu, W.; Wang, J.; Peng, Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer 2020, 6, 319–336.
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691.
- Dumache, R. Early Diagnosis of Oral Squamous Cell Carcinoma by Salivary microRNAs. Clin. Lab. 2017, 63, 1771–1776.
- Liu, Y.; He, A.; Liu, B.; Huang, Z.; Mei, H. Potential Role of lncRNA H19 as a Cancer Biomarker in Human Cancers Detection and Diagnosis: A Pooled Analysis Based on 1585 Subjects. BioMed Res. Int. 2019, 2019, 9056458.
- Luzon-Toro, B.; Fernandez, R.M.; Martos-Martinez, J.M.; Rubio-Manzanares-Dorado, M.; Antinolo, G.; Borrego, S. LncRNA LUCAT1 as a novel prognostic biomarker for patients with papillary thyroid cancer. Sci. Rep. 2019, 9, 14374.
