Engineered bacteriophages (phages) are bacteriophages that have been genetically or chemically altered in some way to create or improve a property for an application, although it would be equally valid to use this term for phages altered for research. Application-directed properties can include: enhanced bacterial killing to improve phage therapy; insertion of reporter genes to facilitate biosensor-mediated detection; insertion of targeting peptides to a virion surface protein to enhance binding properties to bacteria or other types of cells; attachment of non-protein molecules (e.g. antibiotics or nanoparticles) to phage capsid surface proteins to facilitate phage-mediated delivery; anchoring of phages to a surface to improve target capture. It is also possible to combine modifications to develop, for example, a phage-based cancer treatment that has binding peptides for cancer cell targeting and is conjugated to either a radioisotope nanoparticle or chemotherapy drug to improve delivery.
- bacteriophage
- phage therapy
- engineered phage
- imaging agent
1. Introduction
-
Phages are typically very specific in the types of bacterial cells they target
-
Phages are relatively easy and inexpensive to propagate on bacterial hosts using well established protocols
-
Phage capsids are highly stable, often resistant to changes in pH and temperature
-
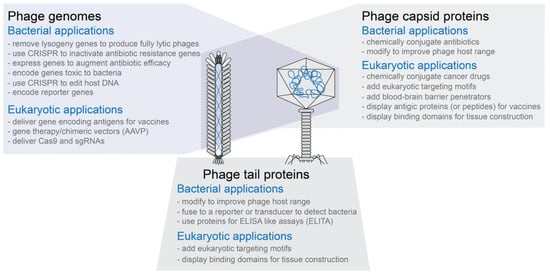
Phage capsids have multiple proteins that can be targeted for modification without necessarily inactivating the phage capsid’s functions (see Figure 1)
-
Phage capsids protect the DNA or RNA packaged in them
-
Phage capsids or modified phage capsids can be used to package non-phage nucleic acid, protein, or other types of materials (see Figure 1)
-
Phage genomes are small compared to bacterial or eukaryotic genomes and are often relatively easy to modify using genetic engineering techniques
-
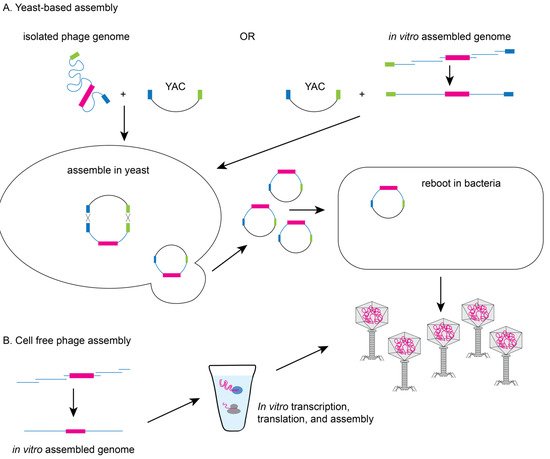
In vitro packaging systems have been created to move genetically modified genomes into capsids as well as transformation protocols to move genomes directly into cells for expression
-
Phage display technology can be used to create phages with novel binding properties even to eukaryotic targets
-
For human treatment applications, phages are generally recognized as safe (GRAS) agents
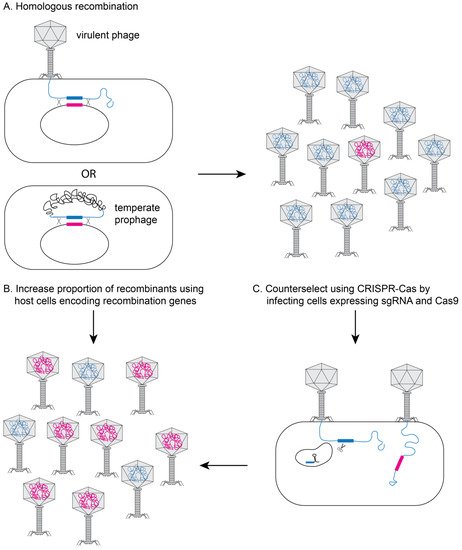
2. Methods to Genetically Engineer Phages
This entry is adapted from the peer-reviewed paper 10.3390/ph14070634
References
- Salmond, G.P.; Fineran, P.C. A century of the phage: Past, present and future. Nat. Rev. Microbiol. 2015, 13, 777–786.
- Letarov, A.V. History of Early Bacteriophage Research and Emergence of Key Concepts in Virology. Biochemistry 2020, 85, 1093–1112.
- Moelling, K.; Broecker, F.; Willy, C. A Wake-Up Call: We Need Phage Therapy Now. Viruses 2018, 10, 688.
- Cooper, C.J.; Mirzaei, M.K.; Nilsson, A.S. Adapting Drug Approval Pathways for Bacteriophage-Based Therapeutics. Front. Microbiol. 2016, 7, 1209.
- Hargreaves, K.R.; Otieno, J.R.; Thanki, A.; Blades, M.J.; Millard, A.D.; Browne, H.P.; Lawley, T.D.; Clokie, M.R. As Clear as Mud? Determining the diversity and prevalence of prophages in the draft genomes of estuarine isolates of clostridium difficile. Genome Biol. Evol. 2015, 7, 1842–1855.
- de Jonge, P.A.; Nóbrega, F.L.; Brouns, S.J.J.; Dutilh, B.E. Molecular and Evolutionary Determinants of Bacteriophage Host Range. Trends Microbiol. 2019, 27, 51–63.
- Pires, D.P.; Cleto, S.; Sillankorva, S.; Azeredo, J.; Lu, T.K. Genetically Engineered Phages: A Review of Advances over the Last Decade. Microbiol. Mol. Biol. Rev. 2016, 80, 523–543.
- Tinoco, J.M.; Buttaro, B.; Zhang, H.; Liss, N.; Sassone, L.M.; Stevens, R. Effect of a genetically engineered bacteriophage on Enterococcus faecalis biofilms. Arch. Oral Biol. 2016, 71, 80–86.
- Kilcher, S.; Loessner, M.J. Engineering Bacteriophages as Versatile Biologics. Trends Microbiol. 2019, 27, 355–367.
- Zhang, H.; Fouts, D.E.; DePew, J.; Stevens, R.H. Genetic modifications to temperate Enterococcus faecalis phage Ef11 that abolish the establishment of lysogeny and sensitivity to repressor, and increase host range and productivity of lytic infection. Microbiology 2013, 159, 1023–1035.
- Ando, H.; Lemire, S.; Pires, D.P.; Lu, T.K. Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst. 2015, 1, 187–196.
- Yosef, I.; Goren, M.G.; Globus, R.; Molshanski-Mor, S.; Qimron, U. Extending the Host Range of Bacteriophage Particles for DNA Transduction. Mol. Cell 2017, 66, 721–728.e3.
- Loessner, M.J.; Rees, C.E.; Stewart, G.S.; Scherer, S. Construction of luciferase reporter bacteriophage A511::luxAB for rapid and sensitive detection of viable Listeria cells. Appl. Environ. Microbiol. 1996, 62, 1133–1140.
- Le, S.; He, X.; Tan, Y.; Huang, G.; Zhang, L.; Lux, R.; Shi, W.; Hu, F. Mapping the Tail Fiber as the Receptor Binding Protein Responsible for Differential Host Specificity of Pseudomonas aeruginosa Bacteriophages PaP1 and JG004. PLoS ONE 2013, 8, e68562.
- Mahichi, F.; Synnott, A.J.; Yamamichi, K.; Osada, T.; Tanji, Y. Site-specific recombination of T2 phage using IP008 long tail fiber genes provides a targeted method for expanding host range while retaining lytic activity. FEMS Microbiol. Lett. 2009, 295, 211–217.
- Sarkis, G.J.; Jacobs, W.R., Jr.; Hatfulll, G.F. L5 luciferase reporter mycobacteriophages: A sensitive tool for the detection and assay of live mycobacteria. Mol. Microbiol. 1995, 15, 1055–1067.
- Tanji, Y.; Furukawa, C.; Na, S.-H.; Hijikata, T.; Miyanaga, K.; Unno, H. Escherichia coli detection by GFP-labeled lysozyme-inactivated T4 bacteriophage. J. Biotechnol. 2004, 114, 11–20.
- Marinelli, L.J.; Hatfull, G.F.; Piuri, M. Recombineering: A powerful tool for modification of bacteriophage genomes. Bacteriophage 2012, 2, 5–14.
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645.
- Fehér, T.; Karcagi, I.; Blattner, F.R.; Pósfai, G. Bacteriophage recombineering in the lytic state using the lambda red recombinases. Microb. Biotechnol. 2012, 5, 466–476.
- Kiro, R.; Shitrit, D.; Qimron, U. Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biol. 2014, 11, 42–44.
- Martel, B.; Moineau, S. CRISPR-Cas: An efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res. 2014, 42, 9504–9513.
- Bari, S.M.N.; Walker, F.C.; Cater, K.; Aslan, B.; Hatoum-Aslan, A. Strategies for Editing Virulent Staphylococcal Phages Using CRISPR-Cas10. ACS Synth. Biol. 2017, 6, 2316–2325.
- Box, A.M.; McGuffie, M.J.; O’Hara, B.J.; Seed, K.D. Functional Analysis of Bacteriophage Immunity through a Type I-E CRISPR-Cas System in Vibrio cholerae and Its Application in Bacteriophage Genome Engineering. J. Bacteriol. 2016, 198, 578–590.
- Lemay, M.-L.; Tremblay, D.M.; Moineau, S. Genome Engineering of Virulent Lactococcal Phages Using CRISPR-Cas9. ACS Synth. Biol. 2017, 6, 1351–1358.
- Schilling, T.; Dietrich, S.; Hoppert, M.; Hertel, R. A CRISPR-Cas9-Based Toolkit for Fast and Precise In Vivo Genetic Engineering of Bacillus subtilis Phages. Viruses 2018, 10, 241.
- Tao, P.; Wu, X.; Tang, W.-C.; Zhu, J.; Rao, V. Engineering of Bacteriophage T4 Genome Using CRISPR-Cas9. ACS Synth. Biol. 2017, 6, 1952–1961.
- Shen, J.; Zhou, J.; Chen, G.-Q.; Xiu, Z.-L. Efficient Genome Engineering of a Virulent Klebsiella Bacteriophage Using CRISPR-Cas9. J. Virol. 2018, 92.
- Jaschke, P.R.; Lieberman, E.K.; Rodriguez, J.; Sierra, A.; Endy, D. A fully decompressed synthetic bacteriophage oX174 genome assembled and archived in yeast. Virology 2012, 434, 278–284.
- Garamella, J.; Marshall, R.; Rustad, M.; Noireaux, V. The All E. coli TX-TL Toolbox 2.0: A Platform for Cell-Free Synthetic Biology. ACS Synth. Biol. 2016, 5, 344–355.
- Shin, J.; Jardine, P.; Noireaux, V. Genome Replication, Synthesis, and Assembly of the Bacteriophage T7 in a Single Cell-Free Reaction. ACS Synth. Biol. 2012, 1, 408–413.
- Rustad, M.; Eastlund, A.; Marshall, R.; Jardine, P.; Noireaux, V. Synthesis of Infectious Bacteriophages in an E. coli-based Cell-free Expression System. J. Vis. Exp. 2017, e56144.
- Rustad, M.; Eastlund, A.; Jardine, P.; Noireaux, V. Cell-free TXTL synthesis of infectious bacteriophage T4 in a single test tube reaction. Synth. Biol. 2018, 3, ysy002.
