Although inhibition of the androgen–androgen receptor (AR) axis effectively represses the growth of prostate cancer, most of all cases eventually become castration-resistant prostate cancers (CRPCs). Enhancement of the expression of AR and its variants along with the downstream signals is important for disease progression. AR-V7, a constitutive active form of AR, is generated as a result of RNA splicing. RNA splicing creates multiple transcript variants from one pre-messenger RNA (mRNA) by removing introns/exons to allow mRNA translation. The molecular mechanisms leading to marked increases of AR and generation of AR-V7 have been unclear. However, recent papers highlighted the roles of RNA splicing factors which promote AR expression and production of variants. Notably, a broad range of splicing components were aberrantly regulated in CRPC tissues. Interestingly, expression of various spliceosome genes is enhanced by RNA-binding protein splicing factor proline- and glutamine-rich (PSF/SFPQ), leading to changes in the expression of AR transcript variants. Moreover, inhibition of several splicing factors repressed tumor growth in vivo. Altered expression of splicing factors is correlated to biochemical recurrence in prostate cancer patients.
1. Introduction
Dysregulation of gene expression is a well-known feature of cancer cells [
1]. Of note, messenger RNA (mRNA) splicing to remove introns from precursor messenger RNA (pre-mRNA) is a critical step in the post-transcriptional regulation of gene expression [
2]. Most human genes generate multiple RNA transcripts through alternative splicing; this phenomenon enables cells to produce a variety of distinct proteins from a single gene [
3]. An enzymatic complex in the nucleus, known as the spliceosome, is responsible for mRNA splicing. The spliceosome consists of five small nuclear ribonucleoproteins, called snRNPs (U1, U2, U4, U5, and U6) [
4,
5,
6,
7,
8,
9]. Each snRNP includes small nuclear RNA (snRNA) complexed to the related proteins, which are named splicing factors. Recent genomic sequencing studies identified that the mutation and aberrant expression of splicing factors are important mechanisms to generate the diversity of gene functions required for cancer progression. Therefore, I present a simplified summary of the spliceosome assembly pathway. In addition, several important reports have demonstrated that the androgen receptor (AR) splicing pathway is a key driver for prostate cancer progression [
10,
11,
12,
13]. Prostate cancer is the second leading cause of cancer deaths in men and the growth of prostate tumors is controlled by the androgen hormone [
14,
15].
2. Splicing Factor Drives Prostate Cancer Progression
The alteration of splicing activity by mutations induces severely abnormal development and tumorigenesis [
3]. However, these mutations have rarely been identified in prostate cancer, although several large scale genomic sequence analyses of primary and metastatic prostate tumors have been conducted [
52]. Previous studies [
52,
53] have identified altered signals in prostate cancer such as TP53, androgen, and Forkhead box protein A1 (FOXA1) signaling, DNA repair, and phosphoinositide3-kinase (PI3K)–AKT signaling. Alternatively, prostate cancer is classified by ETS transcription family fusions or mutations in Speckle-type POZ protein (SPOP). A recent larger study identified that splicing pathway was altered in 4% of prostate tumors [
54]. As observed in other cancer types,
SF3B1 (1.1%) and
U2AF1 (0.5%), which are associated with the recognition of 3’ SS, were mutated. However, the frequency of mutation in these factors is relatively low compared with other types of cancer [
37,
42,
43,
44,
45,
46,
47]. However, several reports have emphasized the importance of
AR splicing in prostate cancer progression [
55]. Altered splicing machinery would result in a dysregulated
AR splicing process.
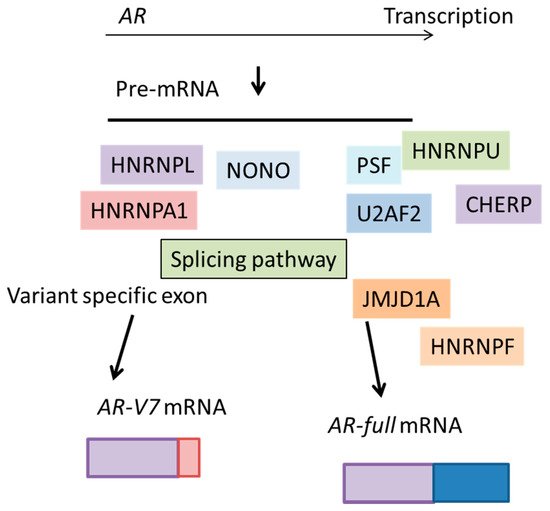
AR-V7 mRNA synthesis requires the removal of intron sequences through an RNA recognition process. The spliceosomal complex excises the 3’ SS and ligates exon 3 with exon 3B by recognizing this exon [
55,
56]. Notably, recent functional and clinical analyses revealed that various splicing factors are highly expressed in CRPC tissue, indicating that the splicing system plays an important role in prostate cancer progression for reasons other than mutation, for example, possibly through overexpression [
22,
57]. The enhanced expression of splicing factors would promote their recruitment to pre-mRNA, facilitating this dynamic and reversible mRNA splicing process. Therefore, I have summarized these representative reports showing several new driving forces for prostate cancer progression and enhanced AR signaling.
2.1. Splicing Factor, Proline- and Glutamine-Rich/ Non-POU Domain-Containing Octamer-Binding Protein (PSF/NONO)
A recent study showed a role of a splicing factor, proline- and glutamine-rich (PSF/SFPQ), in the progression of prostate cancer by using the deep-sequence based approach of clinicopathological analysis [
57]. High PSF protein expression was observed in a subset of tumor samples and higher expression of PSF was associated with cancer-specific survival after surgery and the PSA-free survival after hormone-therapy. In addition,
PSF mRNA expression was significantly elevated in metastatic or advanced prostate cancer samples, suggesting that PSF expression was associated with prostate cancer progression. Using an in vivo model of CRPC, knockdown of PSF reduced the growth of tumors in mice castrated after tumor development. Notably, the study presented a new mechanistic insight, with the identification of global RNAs bound with PSF. Long non-coding (lnc) RNAs, such as androgen-regulated lnc RNA, C-terminal binding protein 1 antisense
(CTBP1-AS) [
58], and a prostate cancer-associated lncRNA, switch/sucrose non-fermentable (SWI/SNF) complex antagonist associated with prostate cancer 1
(SchLAP1) [
59], are positively regulated by PSF in prostate cancer cells. It was also found that various spliceosome genes are uniformly upregulated in metastatic CRPC tissues [
57]. Furthermore, the main targets of PSF at the RNA level are these spliceosome genes (
Figure 3). Widespread upregulation of the splicing pathway in CRPC could affect the splicing complexes in cancer. In addition, by regulating spliceosome gene expressions and cooperating with those factors at protein level, PSF may control the activation a broad range of oncogenic pathways. This study indicates the importance of PSF in the regulation of splicing machinery in aggressive prostate cancer [
57]. These analyses provide an intriguing insight into the mechanism of splicing machinery in cancer progression [
37].
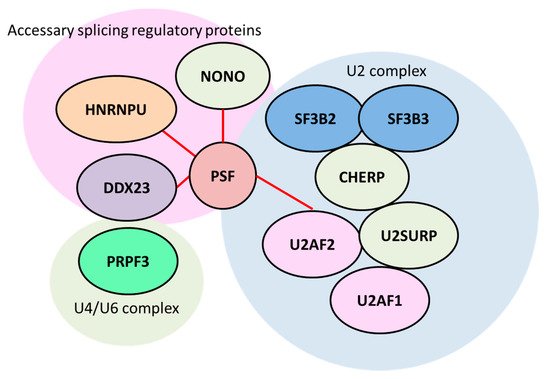
Figure 3. Splicing factor proline- and glutamate-rich (PSF) governs splicing factor complexes in advanced prostate cancer that are involved in AR and cancer-related gene expression. RNA-precipitation and sequencing studies (RIP-seq) revealed that PSF binds to a variety of splicing factors that are uniformly induced in metastatic castration-resistant prostate cancer (CRPC) tissues. The identified PSF-regulated splicing factors that are involved in AR and AR-V7 production are shown. The red line indicates protein–protein interaction validated by immunoprecipitation analysis [
57]. HNRNPU: heterogeneous nuclear ribonucleoprotein U. PRPF3: pre-mRNA processing factor 3. CHERP: calcium homeostasis endoplasmic reticulum protein. DDX23: DEAD (Asp-Glu-Ala-Asp) box (DDX) 23. NONO: non-POU domain-containing octamer-binding protein.
It is notable that one of the most important targets of PSF involved in CRPC is AR, because AR and AR-V7 drives the hormone-refractory state [
55,
56,
60]. Enhanced association of
AR transcript with PSF was observed in CRPC model cells compared with hormone-dependent cancer cells [
58]. PSF regulates
AR splicing process and promotes production of
AR and its variants at the mRNA level. PSF forms a complex with other RNA-binding proteins and non-POU domain-containing octamer-binding protein (NONO) [
61] to promote splicing events of
AR transcripts and AR-V7 production (
Figure 4). Through immunoprecipitation and western blotting analysis, PSF was shown to interact with these spliceosome components, and be responsible for the complex formation of spliceosome. A RIP assay also demonstrated that these factors bound to the intronic region of
AR transcripts and acted as an integrator for the PSF-mediated splicing of
AR transcripts. Thus, PSF orchestrated its target splicing factors at the protein levels to form a complex for splicing and protein expression in CRPC. These functions of RNA-binding proteins could be effective for enhancement of the expressions of prostate cancer-associated genes, such as full-length AR and AR-V7. The expression of these important components, such as CHERP, U2AF2 and HNRNPU [
62,
63,
64,
65,
66], are enhanced by PSF binding. Another report showed that the recruitment of U2AF2 (also called U2AF65) to the 3’ splice site of
AR-V7 with SRSF1 was enhanced by ADT to produce increased expression of AR-V7 [
60]. Collectively, these results suggested that PSF functions as the “commander” of splicing machinery for prostate cancer progression and the AR.
Figure 4. The essential role of various splicing factors for AR-V7 production. Pre-mRNA of AR is spliced by splicing factors upregulated in advanced prostate cancer. In prostate cancer, many splicing factors are transcriptionally induced to modifying the gene expression profile and induce cancer progression. Recently reported representative splicing factors are shown. JMJD1A: Jumonji domain containing 1A. HNRMPL: heterogeneous nuclear ribonucleoprotein L. HNRNPA1: heterogeneous nuclear ribonucleoprotein A1. HNRMPF: heterogeneous nuclear ribonucleoprotein F.
2.2. Heterogeneous Nuclear Ribonucleoprotein (HNRNP) Family Members
A genome-wide clustered regularly interspaced short palindromic repeat (CRISPR) screen has identified the essential genes for prostate cancer growth [
67]. This study demonstrated that heterogeneous nuclear ribonucleoprotein L (HNRNPL) was the most important splicing factor. Comprehensive analysis of HNRNPL-dependent alternative splicing processes in prostate cancer cells identified direct HNRNPL-regulated signals. HNRNPL binds preferentially to cytosine adenine (CA)-repeats or CA-enriched RNA motifs in introns, and the 3’ untranslated region (UTR) regions of genes. Similar to other HNRNP family proteins function in alternative splicing, HNRNPL enhances and represses alternative splicing of different targets [
35]. Overall, the identification of AR as an HNRNPL-regulated splicing target gene at the RNA level is of interest (
Figure 4). As described above, HNRNP family members have important roles as enhancers or repressors of splicing processes [
68,
69]. It has been shown that HNRNP A1 upregulates Lin28 [
70], NKKB2/p52 [
68], and c-Myc [
68], and activates AR-V7. The knockdown of HNRNPA1 downregulated AR-V7 and resensitized enzalutamide-resistant cells to enzalutamide [
68]. In addition,
HNRNPH1 transcripts negatively regulated by
miR-212 were associated with the expression of AR and AR-V7 in prostate cancer cells. HNRNPH1 physically interacts with AR and steroid receptor coactivator-3 (SRC-3), and activates androgen-regulated genes in both a ligand-dependent and -independent manner. Silencing of HNRNPH1 sensitized prostate cancer cells to bicalutamide and inhibited prostate tumor growth in vivo [
71]. These findings suggested that HNRNP family proteins cooperate in the splicing event of AR in CRPC development. Moreover, the HNRNPL-regulated splicing targets were prominently associated with the overexpressed gene signatures in prostate cancer, indicating that these HNRNPL targets may collectively contribute to cancer progression [
67].
HNRNPL is also involved in circular RNA (circRNA) formation [
72,
73]. Trans-factor binding or direct complementary sequence pairing at flanking introns may increase circRNA formation by bringing back spliced exon ends into close proximity [
67]. Notably, the HNRNPL-regulated circRNA genes are relevant to prostate cancer progression, which is suggestive of the potential role of circRNAs produced from these genes [
67].
2.3. Jumonji Domain Containing 1A (JMJD1A)
The histone demethylase, Jumonji domain containing 1A (JMJD1A), was originally found to function as a key coactivator for AR through epigenetic regulation of H3K9 methylation marks [
74,
75]. Moreover, JMJD1A can also act to demethylate non-histone proteins in addition to histone H3K9 [
76,
77]. Another JMJ protein, JMJD6, an arginine demethylase and lysine hydroxylase, can regulate the alternative splicing through the hydroxylation of U2AF2 [
78], suggesting the possibility of JMJ proteins as splicing factors. Indeed, JMJD1A was reported to be involved in the splicing of
AR-V7 in prostate cancer cells [
79] (
Figure 4); mechanistically, HNRNPF was shown to interact with JMJD1A to promote this splicing event [
79]. HNRNPA1 and HNRNPF function together to enhance the splicing events of some pre-mRNAs [
68,
79,
80,
81]. The JMJD1A–HNRNPF complex may interact with HNRNPA1-mediated
AR-V7 splicing. The expression of JMJD1A was significantly associated with that of AR-V7 in prostate cancer tissue. Thus, the level of JMJD1A or factors that regulate the JMJD1A–HNRNPF interaction would promote the alternative splicing of
AR-V7 in prostate cancer. Because increased levels of AR and AR-V7 are important for CRPC progression [
21,
56], current therapeutic strategies aimed at targeting AR are unlikely to have significant effects in prostate cancers expressing ARV7. Therefore, the finding that JMJD1A regulates the activity of AR and the generation of AR-V7 suggests that future development of specific JMJD1A inhibitors should be considered [
79].
This entry is adapted from the peer-reviewed paper 10.3390/biom9040131