Records of eDNA/eRNA exposome may reflect the early appearance, persistence, and presence of biotic and/or abiotic-exposure-mediated modifications in these nucleic acid molecules. Functional genome- and epigenome-wide mapping of eDNA offer great promise to help elucidate the human exposome. Assessment of longitudinal exposure to physical, biological, and chemical agents present in the environment through eDNA/eRNA may enable the building of an integrative causal dynamic stochastic model to estimate environmental causes of human health deficits. Development and validation of monitoring of eDNA/eRNA exposome should seriously be considered to introduce into safety and risk assessment and as surrogates of chronic exposure to environmental stressors.
- eDNA, eRNA, Exposome
1. Definition
The organismal genetic material, deoxyribonucleic acid (DNA), and ribonucleic acid (RNA) present in the environment (home, work or recreational built environment, hazardous wastes, lake and river water, air, etc.) refer to environmental DNA or RNA (eDNA/eRNA). All organisms, including microbes, animals, and humans leave behind traces of genetic material in the environment as they pass through habitats or interact with other organisms. Besides entire microorganism genomes (bacteria, virus, fungus and protozoans), the sources of eDNA/eRNA could be from human or animal blood, hair, excrements (urine, feces, mucous, saliva, secretions), skin, sperm, eggs, roots, leaves, fruit and pollen of plants, and rotting bodies of larger organisms.The eDNA/eRNA-based exposome assessment is expected to rely on our ability to capture the genome- and epigenome-wide signatures left behind by individuals in the indoor and outdoor physical spaces through shedding, excreting, etc.
2. Introduction
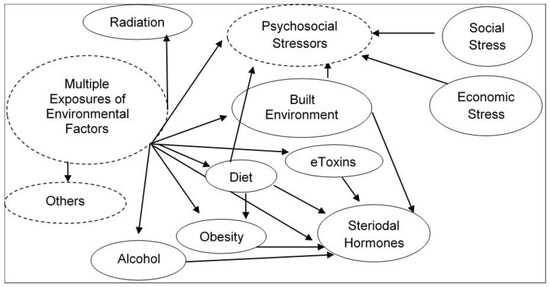
3. eDNA and Environmental Impact Assessment
Hazardous outcomes caused by environmental stressors to ecological health are determined not only by toxic properties, but also changes in the ecosystems, including indigenous biodiversity and community composition. eDNA is one of the most emerging critical biological resources in the field of ecotoxicology [11]. Sequencing and bioinformatic analysis of eDNA samples have been extensively used to comprehensively investigate changes in the ecosystem and use it as a barometer of ecological health [10,12]. Therefore, here we briefly discuss it. Biodiversity monitoring is the standard for environmental impact assessment of anthropogenic activities. eDNA sequencing provides a powerful lens to detect alterations or modifications in biodiversity [13]. High-throughput sequencing (HTS) of eDNA metabarcoding of different ribosomal bacterial and eukaryotic markers is used to measure biotic index values, the taxonomic resolution of molecular markers, and measuring their performance to assess the environmental impact on biodiversity and metabolic function of numerous communities, including soils, ocean, human guts, oral cavity [14,15,16,17]. Most of the ecological and evolutionary metagenomics studies using sequencing of targeted marker genes, such as 16S/18S rDNA/rRNA, offer very limited knowledge about intraspecies diversity and no functional information.
The eDNA has also been used for the detection of rare species, indicator species, or invasive species. This is accomplished by comparing a highly variable region as a barcode in the conserved regions in DNA among species to discriminate between species and determine the taxonomy. To determine various species, metabarcoding eDNA approaches and targeted assays have also been used on ice cores, dirt from swamps, archaeological sites, seawater, fecal samples, and herbal medicines. The application of eDNA metabarcoding extends across many applications. This has provided a huge new tool and more relevant approaches for assessments of risks posed to ecosystems by hazardous agents [11]. Researchers are additionally able to sample organisms and life stages which elude traditional techniques. For example, juvenile and gamete DNA are included in the detection of eDNA from water or sediments. eDNA analysis enables monitoring of the presence of the great crested newt—a protected species in Europe—and detecting invasive bighead carp in the Great Lakes, USA [18,19].
The efforts of studying air pollution are mostly focused on particulate matters in the ambient air, whereas inhalable microorganisms in polluted air have not been well investigated, and the few reported studies mostly focused on taxonomic identification rather than pathogenic potential of microbes. Next-generation sequencing (NGS) of eDNA is not only able to detect invasive species, pathogenic microbes, and responses to pollution, but it can also assess air quality, environmental exposure to chemicals, and their implications in human health [20,21,22,23]. For example, in the case of a major environment insults, such as leaching of toxins from hazardous waste sites, oil spill or intrusion of invasive species, eDNA sequencing and their analysis have been applied to detect what species have been impacted and what genomic changes have occurred in these species over time [24,25,26]. The presence of respiratory microbial allergens and pathogens in outdoors air samples from Beijing collected during a severe smog event and in hospital air samples have been detected by metabarcoding eDNA approaches [27,28]. In summary, these studies provide support, indicating that biodiversity detected by NGS of eDNA can also assess the health risk associated from exposure to environmental agents.
References
- Wild, C.P. The exposome: From concept to utility. Int. J. Epidemiol. 2012, 41, 24–32. [Google Scholar] [CrossRef]
- Deoraj, A.; Yoo, C.; Roy, D. Integrated bioinformatics, biostatistics and molecular epidemiologic approaches to study how the environment and genes work together to influence the development of complex chronic diseases. In Gene Environment Interaction Studies; Anno, S., Ed.; Pan Stanford Publishing: Singapore, 2016; pp. 160–200. [Google Scholar]
- Jansson, J.K.; Hofmockel, K.S. The soil microbiome—From metagenomics to metaphenomics. Curr. Opin. Microbiol. 2018, 43, 162–168. [Google Scholar] [CrossRef]
- Ho, S.Y.W.; Heupink, T.H.; Rambaut, A.; Shapiro, B. Bayesian Estimation of Sequence Damage in Ancient DNA. Mol. Biol. Evol. 2007, 24, 1416–1422. [Google Scholar] [CrossRef]
- Orlando, L.; Cooper, A. Using ancient DNA to understand evolutionary and ecological processes. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 573–598. [Google Scholar] [CrossRef]
- Beng, K.C.; Corlett, R.T. Applications of environmental DNA (eDNA) in ecology and conservation: Opportunities, challenges and prospects. Biodivers. Conserv. 2020, 29, 2089–2121. [Google Scholar] [CrossRef]
- Zaiko, A.; Pochon, X.; Garcia-Vazquez, E.; Olenin, S.; Wood, S.A. Advantages and limitations of environmental DNA/RNA tools for marine biosecurity: Management and surveillance of non-indigenous species. Front. Mar. Sci. 2018, 5, 322. [Google Scholar] [CrossRef]
- Harrison Jori, B.; Sunday Jennifer, M.; Rogers Sean, M. Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proc. R. Soc. B 2019, 286, 20191409. [Google Scholar] [CrossRef]
- Seymour, M. Rapid progression and future of environmental DNA research. Commun Biol. 2019, 2, 80. [Google Scholar] [CrossRef]
- Winding, A.; Bang-Andreasen, T.; Hansen, L.H.; Panitz, F.; Krogh, P.H.; Krause-Jensen, D.; Stæhr, P.; Nicolaisen, M.; Hendriksen, N.B.; Sapkota, R.; et al. eDNA in Environmental Monitoring. Aarhus University, DCE—Danish Centre for Environment and Energy. Technical Report No. 133. 2019. Available online: http://dce2.au.dk/pub/TR133.pdf (accessed on 10 July 2020).
- Zhang, P.; Arora, M.; Chaleckis, R.; Isobe, T.; Jain, M.; Meister, I.; Melén, E.; Perzanowski, M.; Torta, F.; Wenk, M.R.; et al. Tackling the complexity of the exposome: Considerations from the Gunma University Initiative for Advanced Research (GIAR) Exposome Symposium. Metabolites 2019, 9, 106. [Google Scholar] [CrossRef]
- Zinger, L.; Donald, J.; Brosse, S.; Gonzalez, M.A.; Iribar, A.; Leroy, C.; Murienne, J.; Orivel, J.; Schimann, H.; Taberlet, P.; et al. Advances and prospects of environmental DNA in neotropical rainforests. Adv. Ecol. Res. 2020. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 2016, 17, 1–17. [Google Scholar] [CrossRef]
- Mishra, A.; Jha, G.; Thakur, I.S. Draft genome sequence of Zhihengliuella sp. ISTPL4, a psychrotolerant and halotolerant bacterium isolated from Pangong Lake, India. Genome Announc. 2018, 6, e01533-17. [Google Scholar] [CrossRef] [PubMed]
- Rathour, R.; Gupta, J.; Kumar, M.; Hiloidhari, M.; Mehrotra, A.K.; Thakur, I.S. Metagenomic sequencing of microbial communities from a brackish water of pangong lake of north west Indian Himalayas. Genome Announc. 2017, 5, e01029-17. [Google Scholar] [CrossRef]
- Gupta, J.; Rathour, R.; Kumar, M.; Hiloidhari, M.; Mehrotra, A.K.; Thakur, I.S. Metagenomic analysis of microbial diversity in landfill lysimeter soil of Ghazipur landfill site, New Delhi, India. Genome Announc. 2017, 5, e01104. [Google Scholar] [CrossRef]
- Rathour, R.; Gupta, J.; Mishra, A.; Rajeev, A.C.; Dupont, C.L.; Thakur, I.S. A comparative metagenomic study reveals microbial diversity and their role in the biogeochemical cycling of Pangong lake. Sci. Total Environ. 2020. [Google Scholar] [CrossRef]
- Biggs, J.; Ewald, N.; Valentini, A.; Gaboriaud, C.; Dejean, T.; Griffiths, R.A.; Foster, J.; Wilkinson, W.K.; Arnell, A.; Brotherton, P.; et al. Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus). Biol. Conserv. 2015, 183, 19–28. [Google Scholar] [CrossRef]
- Klymus, K.E.; Richter, C.A.; Chapman, D.C.; Paukert, C. Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biol. Conserv. 2014, 183, 77–84. [Google Scholar] [CrossRef]
- Brown, E.A.; Chain, F.J.J.; Zhan, A.; MacIsaac, H.J.; Cristescu, M.E. Early detection of aquatic invaders using metabarcoding reveals a high number of non-indigenous species in Canadian ports. Divers. Distrib. 2016, 22, 1045–1059. [Google Scholar] [CrossRef]
- Zaiko, A.; Martinez, J.L.; Schmidt-Petersen, J.; Ribicic, D.; Samuiloviene, A.; Garcia-Vazquez, E. Metabarcoding approach for the ballast water surveillance an advantageous solution or an awkward challenge? Mar. Pollut. Bull. 2015, 92, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Zaiko, A.; Samuiloviene, A.; Ardura, A.; Garcia-Vazquez, E. Metabarcoding approach for nonindigenous species surveillance in marine coastal waters. Mar. Pollut. Bull. 2015, 100, 53–59. [Google Scholar] [CrossRef]
- Zaiko, A.; Schimanski, K.; Pochon, X.; Hopkins, G.A.; Goldstien, S.; Floerl, O.; Wood, S.A. Metabarcoding improves detection of eukaryotes from early biofouling communities: Implications for pest monitoring and pathway management. Biofouling J. Bioadh. Biofilm Res. 2016, 32, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Lanzen, A.; Lekang, K.; Jonassen, I.; Thompson, E.M.; Troedsson, C. High-throughput metabarcoding of eukaryotic diversity for environmental monitoring of offshore oil-drilling activities. Mol. Ecol. 2016, 25, 4392–4406. [Google Scholar] [CrossRef] [PubMed]
- Lanzen, A.; Lekang, K.; Jonassen, I.; Thompson, E.M.; Troedsson, C. DNA extraction replicates improve diversity and compositional dissimilarity y in metabarcoding of eukaryotes in marine sediments. PLoS ONE 2017, 12, e0179443. [Google Scholar] [CrossRef] [PubMed]
- Laroche, O.; Wood, S.A.; Tremblay, L.A.; Ellis, J.I.; Lear, G.; Pochon, X. A cross-taxa study using environmental DNA/RNA metabarcoding to measure biological impacts of offshore oil and gas drilling and production operations. Mar. Pollut. Bull. 2018, 127, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Jiang, W.; Wang, B.; Fang, J.; Lang, J.; Tian, G.; Jiang, J.; Zhu, T.F. Inhalable microorganisms in Beijing’s PM2.5 and PM10 pollutants during a severe smog event. Environ. Sci. Technol. 2014, 48, 1499–14507. [Google Scholar] [CrossRef]
- Tong, X.; Xu, H.; Zou, L.; Cai, M.; Xu, X.; Zhao, Z.; Xiao, F.; Li, Y. High diversity of airborne fungi in the hospital environment as revealed by metasequencing-based microbiome analysis. Sci. Rep. 2017, 7, 39606. [Google Scholar] [CrossRef]
- Wild, C.P. The exposome: From concept to utility. Int. J. Epidemiol. 2012, 41, 24–32. [Google Scholar] [CrossRef]
- Deoraj, A.; Yoo, C.; Roy, D. Integrated bioinformatics, biostatistics and molecular epidemiologic approaches to study how the environment and genes work together to influence the development of complex chronic diseases. In Gene Environment Interaction Studies; Anno, S., Ed.; Pan Stanford Publishing: Singapore, 2016; pp. 160–200. [Google Scholar]
- Jansson, J.K.; Hofmockel, K.S. The soil microbiome—From metagenomics to metaphenomics. Curr. Opin. Microbiol. 2018, 43, 162–168. [Google Scholar] [CrossRef]
- Ho, S.Y.W.; Heupink, T.H.; Rambaut, A.; Shapiro, B. Bayesian Estimation of Sequence Damage in Ancient DNA. Mol. Biol. Evol. 2007, 24, 1416–1422. [Google Scholar] [CrossRef]
- Orlando, L.; Cooper, A. Using ancient DNA to understand evolutionary and ecological processes. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 573–598. [Google Scholar] [CrossRef]
- Beng, K.C.; Corlett, R.T. Applications of environmental DNA (eDNA) in ecology and conservation: Opportunities, challenges and prospects. Biodivers. Conserv. 2020, 29, 2089–2121. [Google Scholar] [CrossRef]
- Zaiko, A.; Pochon, X.; Garcia-Vazquez, E.; Olenin, S.; Wood, S.A. Advantages and limitations of environmental DNA/RNA tools for marine biosecurity: Management and surveillance of non-indigenous species. Front. Mar. Sci. 2018, 5, 322. [Google Scholar] [CrossRef]
- Harrison Jori, B.; Sunday Jennifer, M.; Rogers Sean, M. Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proc. R. Soc. B 2019, 286, 20191409. [Google Scholar] [CrossRef]
- Seymour, M. Rapid progression and future of environmental DNA research. Commun Biol. 2019, 2, 80. [Google Scholar] [CrossRef]
- Winding, A.; Bang-Andreasen, T.; Hansen, L.H.; Panitz, F.; Krogh, P.H.; Krause-Jensen, D.; Stæhr, P.; Nicolaisen, M.; Hendriksen, N.B.; Sapkota, R.; et al. eDNA in Environmental Monitoring. Aarhus University, DCE—Danish Centre for Environment and Energy. Technical Report No. 133. 2019. Available online: http://dce2.au.dk/pub/TR133.pdf (accessed on 10 July 2020).
- Zhang, P.; Arora, M.; Chaleckis, R.; Isobe, T.; Jain, M.; Meister, I.; Melén, E.; Perzanowski, M.; Torta, F.; Wenk, M.R.; et al. Tackling the complexity of the exposome: Considerations from the Gunma University Initiative for Advanced Research (GIAR) Exposome Symposium. Metabolites 2019, 9, 106. [Google Scholar] [CrossRef]
- Zinger, L.; Donald, J.; Brosse, S.; Gonzalez, M.A.; Iribar, A.; Leroy, C.; Murienne, J.; Orivel, J.; Schimann, H.; Taberlet, P.; et al. Advances and prospects of environmental DNA in neotropical rainforests. Adv. Ecol. Res. 2020. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 2016, 17, 1–17. [Google Scholar] [CrossRef]
- Mishra, A.; Jha, G.; Thakur, I.S. Draft genome sequence of Zhihengliuella sp. ISTPL4, a psychrotolerant and halotolerant bacterium isolated from Pangong Lake, India. Genome Announc. 2018, 6, e01533-17. [Google Scholar] [CrossRef] [PubMed]
- Rathour, R.; Gupta, J.; Kumar, M.; Hiloidhari, M.; Mehrotra, A.K.; Thakur, I.S. Metagenomic sequencing of microbial communities from a brackish water of pangong lake of north west Indian Himalayas. Genome Announc. 2017, 5, e01029-17. [Google Scholar] [CrossRef]
- Gupta, J.; Rathour, R.; Kumar, M.; Hiloidhari, M.; Mehrotra, A.K.; Thakur, I.S. Metagenomic analysis of microbial diversity in landfill lysimeter soil of Ghazipur landfill site, New Delhi, India. Genome Announc. 2017, 5, e01104. [Google Scholar] [CrossRef]
- Rathour, R.; Gupta, J.; Mishra, A.; Rajeev, A.C.; Dupont, C.L.; Thakur, I.S. A comparative metagenomic study reveals microbial diversity and their role in the biogeochemical cycling of Pangong lake. Sci. Total Environ. 2020. [Google Scholar] [CrossRef]
- Biggs, J.; Ewald, N.; Valentini, A.; Gaboriaud, C.; Dejean, T.; Griffiths, R.A.; Foster, J.; Wilkinson, W.K.; Arnell, A.; Brotherton, P.; et al. Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus). Biol. Conserv. 2015, 183, 19–28. [Google Scholar] [CrossRef]
- Klymus, K.E.; Richter, C.A.; Chapman, D.C.; Paukert, C. Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biol. Conserv. 2014, 183, 77–84. [Google Scholar] [CrossRef]
- Brown, E.A.; Chain, F.J.J.; Zhan, A.; MacIsaac, H.J.; Cristescu, M.E. Early detection of aquatic invaders using metabarcoding reveals a high number of non-indigenous species in Canadian ports. Divers. Distrib. 2016, 22, 1045–1059. [Google Scholar] [CrossRef]
- Zaiko, A.; Martinez, J.L.; Schmidt-Petersen, J.; Ribicic, D.; Samuiloviene, A.; Garcia-Vazquez, E. Metabarcoding approach for the ballast water surveillance an advantageous solution or an awkward challenge? Mar. Pollut. Bull. 2015, 92, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Zaiko, A.; Samuiloviene, A.; Ardura, A.; Garcia-Vazquez, E. Metabarcoding approach for nonindigenous species surveillance in marine coastal waters. Mar. Pollut. Bull. 2015, 100, 53–59. [Google Scholar] [CrossRef]
- Zaiko, A.; Schimanski, K.; Pochon, X.; Hopkins, G.A.; Goldstien, S.; Floerl, O.; Wood, S.A. Metabarcoding improves detection of eukaryotes from early biofouling communities: Implications for pest monitoring and pathway management. Biofouling J. Bioadh. Biofilm Res. 2016, 32, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Lanzen, A.; Lekang, K.; Jonassen, I.; Thompson, E.M.; Troedsson, C. High-throughput metabarcoding of eukaryotic diversity for environmental monitoring of offshore oil-drilling activities. Mol. Ecol. 2016, 25, 4392–4406. [Google Scholar] [CrossRef] [PubMed]
- Lanzen, A.; Lekang, K.; Jonassen, I.; Thompson, E.M.; Troedsson, C. DNA extraction replicates improve diversity and compositional dissimilarity y in metabarcoding of eukaryotes in marine sediments. PLoS ONE 2017, 12, e0179443. [Google Scholar] [CrossRef] [PubMed]
- Laroche, O.; Wood, S.A.; Tremblay, L.A.; Ellis, J.I.; Lear, G.; Pochon, X. A cross-taxa study using environmental DNA/RNA metabarcoding to measure biological impacts of offshore oil and gas drilling and production operations. Mar. Pollut. Bull. 2018, 127, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Jiang, W.; Wang, B.; Fang, J.; Lang, J.; Tian, G.; Jiang, J.; Zhu, T.F. Inhalable microorganisms in Beijing’s PM2.5 and PM10 pollutants during a severe smog event. Environ. Sci. Technol. 2014, 48, 1499–14507. [Google Scholar] [CrossRef]
- Tong, X.; Xu, H.; Zou, L.; Cai, M.; Xu, X.; Zhao, Z.; Xiao, F.; Li, Y. High diversity of airborne fungi in the hospital environment as revealed by metasequencing-based microbiome analysis. Sci. Rep. 2017, 7, 39606. [Google Scholar] [CrossRef]
This entry is adapted from the peer-reviewed paper 10.3390/ijms21144879
