Recent data point to the leading role of heterochromatin in genome maintenance, highlighting the attractions between heterochromatic regions as being central to phase separation of the active and inactive chromatin domains. The three main properties of heterochromatin—the position silencing effect on transcription, stickiness, and flexible rigidity—are important for its network formation.
- chromatin organization
- heterochromatin
- networks
- positional information
- scale-free oscillations
- nucleolar boundary
- transcriptional pulsing
- cytoskeleton
- physics of life
1. Introduction
The DNA linear sequence code represents only the most basic principle of gene regulation. In 1944, Erwin Schrödinger (one of the founders of quantum mechanics and Physics Nobel Prize winner of 1933) published his famous lecture “What is life?”. He proposed that DNA may possess the features of aperiodic crystal and use the spatial organization as one of the regulatory elements. The physical features of heterochromatin composed of repeated short nucleotide sequences (satellite DNA) may confer the specificity to provide a gene with “a chromosomal address” [1]. In turn, Ilya Prigogine, another Nobel prize winner in chemistry 1977, formulated that any living dissipative system undergoing exchange of matter and energy with the environment is a subject of structured attenuated oscillations[2][3][4]. Thus, the 3D nuclear organization experiences transcriptional oscillations coordinating its function and scaling many orders of magnitude, including cell-cell interaction [5][6][7][8][9]. At a very coarse grain, we can imagine such oscillations as a biphasic interchange of chromatin compartments between active euchromatin and inactive ( heterochromatin compartment [10][11]. Although the dominating view in the last decade was that the euchromatin and heterochromatin compartments behave as relatively independent self-organized crowded fluids, some recent data still point to the leading role of heterochromatin in genome maintenance, highlighting the attractions between heterochromatic regions as being central to phase separation of the active and inactive chromatin domains [12][13] Recent experiments have shown that heterochromatin and bound HP1 protein undergo liquid-liquid phase separation, while euchromatin does not [14][15][16]. In [17] it was revealed that condensed chromatin exists in a solid-like state whose properties resist external forces and create an elastic gel providing a scaffold that supports liquid-liquid phase separation of chromatin-binding proteins. The physical features of constitutive heterochromatin, namely the regular structure, condensation, and positioning of nucleosomes, are conformed by a set of associated chromosomal proteins, including subcompartmentation by CTCF and cohesin, [15][18][19] which also depend evolutionarily on condensin II[20].
The properties of heterochromatin are consistent with the earlier ideas that its topology, in terms of the sum of its position-cis-silencing effects on transcription, can provide the 3D “genome active space” in addition to the DNA gene-coding function, “the morphogenic function” [21]—a source of positional information necessary for the whole genome regulation [8][22][23][24][25][26][27][28]. In the recent publication [29], researchers were able to link the timing of structural bursting of pericentromere-associated domains (PADs) with the large-scale activation of gene expression occurring by critical self-organization in cancer cells coinciding with biphasic activation of bivalent stress-response genes of the FOS family in the differentiation commitment model. Loss of the heterochromatin silencing threshold by splitting PAD clusters leading to the documented massive unfolding of euchromatin was suggested as the first step for genome repatterning. The discovered basic features of PADs, converting the same amount of constitutive heterochromatin by fusing-splitting within the constraint of cell nucleus volume, suggests their involvement in the supra-chromosomal network as one of the most promising structural candidates supporting chromatin driving gradients and transcriptional pulsing by bursting synchronization[30].
The mesoscopic approach to the description of complex systems is guided by unveiling the presence of “organization principles”. Its main peculiarity is the independence from the detailed knowledge of the basic bricks of the system and the emergence of relevant features from the correlation structure of the studied phenomenon, where “all the entities can be considered as networks of interacting parts” [31]. Here, we will follow this approach to individuate (besides a detailed molecular-level mechanistic approach) some basic pillars of the supra-chromosome wiring structure.
2. Flexible Heterochromatin, Transcription Regulation, and Related Mechanobiology of the Actomyosin Network
The mechanobiology of transcription and transcriptional pulsing itself. EM studies of speckles (also called interchromatin granule clusters) examined in very active cancer cells revealed their direct link to both perinucleolar and perinuclear heterochromatin. Suppression of rRNA synthesis by AcD provoked the radial-concentric organization of the main components involved in transcription machinery, with heterochromatin disclosing a spatial link between rRNA and mRNA synthesis and processing events. The situation with low AcD mimics a return phase of transcriptional pulsing.
An actomyosin-based cytoskeleton using ATP energy plays a role in heterochromatin segregation and the organization of specific heterochromatin compartments: β-actin deficient cells exhibit changes in the spatial organization of H3K9Me3/HP1α—positive heterochromatin[32]. At the same time, one of the primary functions of the elastic nuclear actomyosin network is to maintain a chromatin landscape compatible with transcription [32][33][34]. How are these two functions coupled through the putative nuclear heterochromatin network?
The involvement of fibrillar nuclear actin (F-actin) in the activation and process of transcription with all three RNA polymerases is well-established [33][35][36][37][38]. Upon serum stimulation of transcription, F-actin is seen accumulating in the nucleolus, transcription fabrics, and at the nuclear envelope [39] outlining the route of the transcription conveyor. Nuclear actin and the motor of actin, nuclear myosin (NM1), are required for RNA polymerase I transcription of rRNA [37][40]. Under AcD action, NM1 is transferred into the nucleolus [41], likely favouring the contraction of NORs.
Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II; it is involved both in the initiation and elongation of transcription, acting through ribonucleoprotein interactions and the chromatin remodelling, but also interacting with pure DNA [42][43]. A/B-type proteins [44][45]. NM1-F-actin polymerizing complex activating Pol II needs the binding of bipolar phosphatidylinositol 4,5-bisphosphate and adapting by its disordered protein domain, which also enables liquid phase separation of nuclear speckles [46]. The role of speckles in the amplification of gene expression was recently stressed [47].
The involvement in RNA transcription of actomyosin activity of nuclear speckles, their phase separation, their anchoring to the perinucleolar and perinuclear heterochromatin, as well as their spring-like morphology, visualized by EM (Figure 8a,b), is apparent. NM1 interacts with rRNA transcription initiation factor TIF-IA, which brings actin and NM1 in close proximity with each other. It thus provides feedback with the nucleolus sensing the cell environment, starting transcription pulsing aided by actomyosin. These studies show that the regulation of transcription is structured.
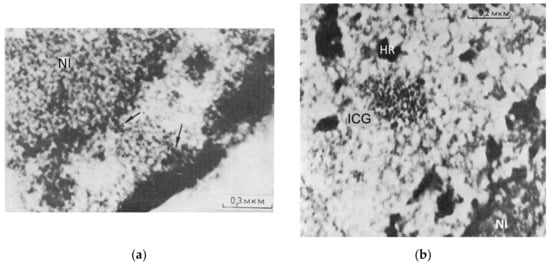
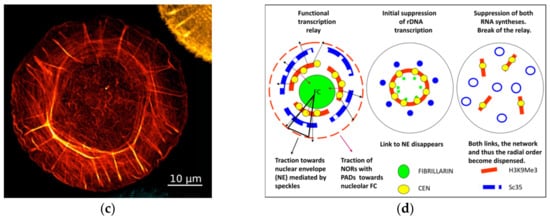
Figure 1. (c) The representative image of the circular-radial self-organization of F-actin filaments (stained by phalloidin) of human foreskin fibroblasts, structured soon after seeding on an isotropic substrate; from [48]. (d) Schematic of the functional transcriptional relay in its relationship with speckles and the concentric rings of the nucleolar and perinuclear heterochromatin shells, deduced from experiments with suppression of RNA synthesis and the literature on the participation of the nuclear cytoskeleton in it. Modified from [49].
It presumes that radial nuclear order is much dependent on the speckle springy traction of the nucleolar and nuclear RNA synthesis conveyor between the perinucleolar and perinuclear heterochromatin stiffly bending concentric rings. Release of the connection to nuclear lamin contracts the system inside the cell nucleus and exposes its radial-concentric character. When nuclear synthesis has been fully suppressed, both anchors of speckles to the perinucleolar and perinuclear constraints are released and the radial-concentric order is dispensed. Thus, nuclear speckles postulated as a structure integrating the whole process of transcription [50][51], seem indeed capable of executing it through transcription pulsing of the heterochromatin network united with the actomyosin network which, in its turn, also follows and supports its radial-concentric character.
3. Nucleolar Pulsing and Nucleolus-Associated Domains (NADs)
Balbiani was the first who discovered vacuoles and their discharge from the nucleolus in the active cells and called the nucleolus the “heart of the cell” [52]. Nucleolar pulsing was also described in plants, associated with transcriptional activity [53][54] and in mouse oocytes with the particularly high synthesis of rRNA [55]. Pulsation of nucleolar rRNA synthesis was also revealed using confocal Raman microspectroscopy [56], and pulsation and coalescence of human nucleoli were registered by [57]. The authors suggest that observation of discontinuous transcriptional activity in the nucleolus may reflect cycling in the assembly and disassembly of active chromatin structure in and/or around rDNA genes.
In this connection, the question arises whether the emergence of function in transcription alveoli may be associated with nucleolus-associated domains (NADs). rDNAs are the most abundant genes, and the stability of the rDNA cluster was considered a key element of genome maintenance [58].
NADs are defined as genomic regions enriched at the nucleolar periphery in interphase cells [59]. In addition to the nucleolar organizers (NORs) in five acrocentric chromosome pairs of the human genome [60], when using deep sequencing of chromatin associated with biochemically purified nucleoli in Hi-C and other labelling methods, it was revealed that specific chromatin domains from most human chromosomes may associate with nucleoli [61]. The NADs comprising 38%
of the human genome contain rDNA sequences and H3K9me3-enriched heterochromatin associated with a-satellite DNA and centromeres [62]. The most recent data on NADs in mouse embryonic stem cells and fibroblasts, besides a large NADs subset overlapping with LADs, revealed a subset of smaller NADs free from binding to nuclear lamin—corresponding more dynamic chromocenters with higher gene density, including developmental and differentiation genes [63][64]. Nucleoli in G1 cells are very dynamic—fusing and splitting; the reiterated ribosomal DNA sequences are also easily amplified and highly variable in size [65][66][67], flexibly adapting to cell needs, and are the first undergoing replication stress during genome instability and cell senescence [68].
In any case, the available molecular and morphological data indicate that most NADs are associated with PADs, and this link can be hypothesized to play a role in the formation of functional alveoli and scale-free transcriptional pulsing of their mutual network coordinating rRNA and mRNA synthesis in the radially directed relay. Therefore, we should shortly review some data on transcriptional pulsing.
4. Transcriptional Pulsing and Synchronization in a Cell Population
Transcriptional bursting, also known as transcriptional pulsing, is a fundamental property of genes, which have been observed in diverse organisms, from bacteria to mammals [69]. The burst frequency is primarily encoded in enhancers, and burst size in core promoters, while cell type-specific gene expression is primarily shaped by changes in burst frequencies [12]. Cultured cells display cell cycle-independent collective dynamics of gene transcription fluctuations with a period of 20–40 min [9]. The stress response of Zaidela ascites carcinoma cells (washed and incubated in PBS in a plugged vial), seen as a few ~90 min collective waves of mRNA synthesis increased up to two to three times, was registered along with a bi-stable switch for the increased superhelicity of DNA opposed by the emergence and subsequent repair of DNA single-strand-breaks; these oscillations further faded and led to cell death [13][70].
The sense of radial-circular nuclear organization for transcriptional pulsing is not only in the structural conveyor coordinating the production of pre-ribosomes with the synthesis and maturation of It means feedback from sensing the environment and cell community which, in case of life threat (and likely, in reprogramming), can become synchronised. In other words—it is an element of explorative adaptation requiring the system to act as a whole. But the question remains, how does this dynamicity fit with the robust regulation of gene expression in tissue differentiation [71] and, ultimately, how is the tissue-specific code established and read?
This entry is adapted from the peer-reviewed paper 10.3390/cells10071582
References
- Smirnov, E.; Hornáček, M.; Vacík, T.; Cmarko, D.; Raška, I. Discontinuous Transcription. Nucleus 2018, 9, 149–160.
- Larsson, A.J.M.; Johnsson, P.; Hagemann-Jensen, M.; Hartmanis, L.; Faridani, O.R.; Reinius, B.; Segerstolpe, Å.; Rivera, C.M.; Ren, B.; Sandberg, R. Genomic Encoding of Transcriptional Burst Kinetics. Nature 2019, 565, 251–254.
- Tsuchiya, M.; Wong, S.T.; Yeo, Z.X.; Colosimo, A.; Palumbo, M.C.; Farina, L.; Crescenzi, M.; Mazzola, A.; Negri, R.; Bianchi, M.M.; et al. Gene Expression Waves. Cell Cycle Independent Collective Dynamics in Cultured Cells. FEBS J. 2007, 274, 2878–2886.
- Erenpreisa, J.; Budylin, A. Related Changes in RNA Synthesis and DNA Superhelicity during Starvation of Ehrlich Ascites Tumour Cells. Proc. Latv. Acad. Sci. Ser. B 1990, 10, 90–94.
- Budylin, A.; Erenpreisa, J.; Zhukotsky, A.; Zaleskaya, N. Fluctuations of the Reparable DNA Strand Breaks in Starving Tumour Cells. Proc. Latv. Acad. Sci. 1993, 5, 72–73.
- Tsuchiya, M.; Giuliani, A.; Hashimoto, M.; Erenpreisa, J.; Yoshikawa, K. Self-Organizing Global Gene Expression Regulated through Criticality: Mechanism of the Cell-Fate Change. PLoS ONE 2016, 11, e0167912.
- Bizhanova, A.; Yan, A.; Yu, J.; Zhu, L.J.; Kaufman, P.D. Distinct Features of Nucleolus-Associated Domains in Mouse Embryonic Stem Cells. Chromosoma 2020, 129, 121–139.
- Caragine, C.M.; Haley, S.C.; Zidovska, A. Nucleolar Dynamics and Interactions with Nucleoplasm in Living Cells. Elife 2019, 8, e47533.
- Wong, L.H.; Brettingham-Moore, K.H.; Chan, L.; Quach, J.M.; Anderson, M.A.; Northrop, E.L.; Hannan, R.; Saffery, R.; Shaw, M.L.; Williams, E.; et al. Centromere RNA Is a Key Component for the Assembly of Nucleoproteins at the Nucleolus and Centromere. Genome Res. 2007, 17, 1146–1160.
- Porokhovnik, L.N.; Lyapunova, N.A. Dosage Effects of Human Ribosomal Genes (rDNA) in Health and Disease. Chromosome Res. 2019, 27, 5–17.
- Roberts, P.; Moshitch-Moshkovitz, S.; Kvam, E.; O’Toole, E.; Winey, M.; Goldfarb, D.S. Piecemeal Microautophagy of Nucleus in Saccharomyces Cerevisiae. Mol. Biol. Cell 2003, 14, 129–141.
- Giuliani, A. The Statistical Mechanics of Life: Comment on “Dynamic and Thermodynamic Models of Adaptation” by A.N. Gorban et al. Phys. Life Rev. 2021, 37, 100–102.
- Xie, X.; Percipalle, P. An Actin-Based Nucleoskeleton Involved in Gene Regulation and Genome Organization. Biochem. Biophys. Res. Commun. 2018, 506, 378–386.
- de Lanerolle, P. Nuclear Actin and Myosins at a Glance. J. Cell Sci. 2012, 125, 4945–4949.
- Venit, T.; Mahmood, S.R.; Endara-Coll, M.; Percipalle, P. Nuclear Actin and Myosin in Chromatin Regulation and Maintenance of Genome Integrity. Int. Rev. Cell Mol. Biol. 2020, 355, 67–108.
- Hofmann, W.A.; Vargas, G.M.; Ramchandran, R.; Stojiljkovic, L.; Goodrich, J.A.; de Lanerolle, P. Nuclear Myosin I Is Necessary for the Formation of the First Phosphodiester Bond during Transcription Initiation by RNA Polymerase II. J. Cell. Biochem. 2006, 99, 1001–1009.
- Hu, P.; Wu, S.; Hernandez, N. A Role for Beta-Actin in RNA Polymerase III Transcription. Genes Dev. 2004, 18, 3010–3015.
- Philimonenko, V.V.; Zhao, J.; Iben, S.; Dingová, H.; Kyselá, K.; Kahle, M.; Zentgraf, H.; Hofmann, W.A.; de Lanerolle, P.; Hozák, P.; et al. Nuclear Actin and Myosin I Are Required for RNA Polymerase I Transcription. Nat. Cell Biol. 2004, 6, 1165–1172.
- Yamazaki, S.; Yamamoto, K.; Harata, M. Contribution of Nuclear Actin to Transcription Regulation. Genom Data 2015, 4, 127–129.
- Wei, M.; Fan, X.; Ding, M.; Li, R.; Shao, S.; Hou, Y.; Meng, S.; Tang, F.; Li, C.; Sun, Y. Nuclear Actin Regulates Inducible Transcription by Enhancing RNA Polymerase II Clustering. Sci. Adv. 2020, 6, eaay6515.
- Ye, J.; Zhao, J.; Hoffmann-Rohrer, U.; Grummt, I. Nuclear Myosin I Acts in Concert with Polymeric Actin to Drive RNA Polymerase I Transcription. Genes Dev. 2008, 22, 322–330.
- Pranchevicius, M.C.S.; Baqui, M.M.A.; Ishikawa-Ankerhold, H.C.; Lourenço, E.V.; Leão, R.M.; Banzi, S.R.; dos Santos, C.T.; Roque-Barreira, M.C.; Espreafico, E.M.; Larson, R.E. Myosin Va Phosphorylated on Ser1650 Is Found in Nuclear Speckles and Redistributes to Nucleoli upon Inhibition of Transcription. Cell Motil. Cytoskelet. 2008, 65, 441–456.
