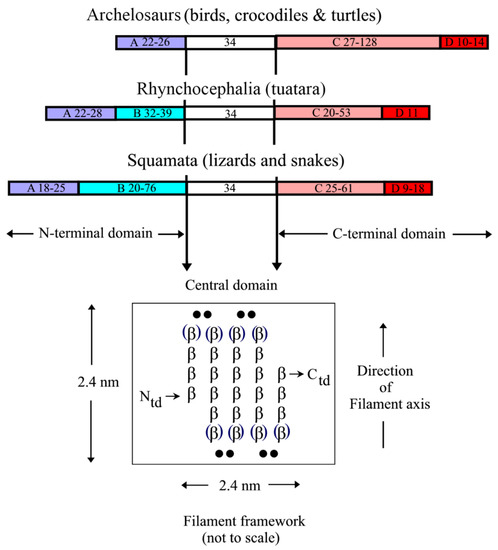
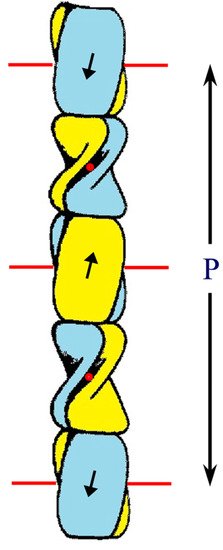
The epidermal appendages of birds and reptiles (the sauropsids) include claws, scales, and feathers. Each has specialized physical properties that facilitate movement, thermal insulation, defence mechanisms, and/or the catching of prey. The mechanical attributes of each of these appendages originate from its fibril-matrix texture, where the two filamentous structures present, i.e., the corneous ß-proteins (CBP or ß-keratins) that form 3.4 nm diameter filaments and the α-fibrous molecules that form the 7–10 nm diameter keratin intermediate filaments (KIF), provide much of the required tensile properties. The matrix, which is composed of the terminal domains of the KIF molecules and the proteins of the epidermal differentiation complex (EDC) (and which include the terminal domains of the CBP), provides the appendages, with their ability to resist compression and torsion.
- corneous ß-proteins
- keratin intermediate filaments
- X-ray fiber diffraction
1. Introduction
2. Structure of the Corneous ß-Protein Molecules and Their Assembly into Filaments
This entry is adapted from the peer-reviewed paper 10.3390/genes12040591
References
- Astbury, W.T.; Marwick, T.C. X-ray interpretation of the molecular structure of feather keratin. Nature 1932, 130, 309–310.
- Fraser, R.D.B.; MacRae, T.P. Molecular organization in feather keratin. J. Mol. Biol. 1959, 1, 387–397.
- Fraser, R.D.B.; MacRae, T.P. Structural organization in feather keratin. J. Mol. Biol. 1963, 7, 272–280.
- Schorr, R.; Krimm, S. Studies on the structure of feather keratin I. X-ray diffraction studies and other experimental data. Biophys. J. 1961, 1, 467–487.
- Stewart, M. The structure of chicken scale keratin. J. Ultrastruct. Res. 1977, 60, 27–33.
- Rudall, K.M. X-ray studies of the distribution of protein chain types in the vertebrate epidermis. Biochim. Biophys. Acta 1947, 1, 549–562.
- O’Donnell, I.J. The complete amino acid sequence of a feather keratin from emu (Dromaius novae-hollandiae). Aust. J. Biol. Sci. 1973, 26, 415–437.
- Fraser, R.D.B.; Suzuki, E. Polypeptide chain conformation in feather keratin. J. Mol. Biol. 1965, 14, 279–282.
- Suzuki, E. Localization of beta-conformation in feather keratin. Aust. J. Biol. Sci. 1973, 26, 435–437.
- Fraser, R.D.B.; MacRae, T.P. The molecular structure of feather keratin. In Proceedings of the 16th International Ornithological Congress, Canberra, Australia, 12–17 August 1976; pp. 443–451.
- Sawyer, R.H.; Glenn, T.; French, J.O.; Mays, B.; Shames, R.B.; Barnes, G.L.; Rhodes, W.; Ishikawa, Y. The expression of beta (β) keratins in the epidermal appendages of reptiles and birds. Am. Zool. 2000, 40, 530–539.
- Alibardi, L.; Toni, M. β-keratins of reptilian scales share a central amino acid sequence termed core-box. Res. J. Biol. Sci. 2007, 2, 329–339.
- Dalla Valle, L.; Nardi, G.B.; Bonazza, G.; Zuccal, C.; Emera, D.; Alibardi, L. Forty keratin-associated β-proteins (β-proteins) form the hard layers of scales, claws, adhesive pads in the green anole lizard, Anolis carolinensis. J. Exp. Zool. Part B Mol. Dev. Evol. 2010, 314B, 11–32.
- Inglis, A.S.; Gillespie, M.J.; Roxborough, C.M.; Whitaker, L.A.; Casagranda, F. Sequence of a glycine-rich protein from lizard claw: Unusual dilute acid and heptafluorobutyric acid cleavages. In Proteins, Structure and Function; L’Italien, J.L., Ed.; Plenum: New York, NT, USA, 1987; pp. 757–764.
- Fraser, R.D.B.; Parry, D.A.D. The molecular structure of reptilian keratin. Int. J. Biol. Macromol. 1996, 19, 207–211.
- Fraser, R.D.B.; Parry, D.A.D. Amino acid sequence homologies in the hard keratins of birds and reptiles, and their implications for molecular structure and physical properties. J. Struct. Biol. 2014, 188, 213–224.
- Fraser, R.D.B.; Parry, D.A.D. The structural basis of the filament-matrix texture in the avian/reptilian group of hard β-keratins. J. Struct. Biol. 2011, 173, 391–405.
- Fraser, R.D.B.; Parry, D.A.D. Molecular packing in the feather keratin filament. J. Struct. Biol. 2008, 162, 1–13.
- Fraser, R.D.B.; Parry, D.A.D. Filamentous structure of hard β-keratin in the epidermal appendages of birds and reptiles. In Subcellular Biochemistry: Fibrous Proteins: Structures and Mechanisms; Parry, D.A.D., Squire, J.M., Eds.; Springer: Cham, Switzerland, 2017; Volume 82, pp. 231–252.
- Chothia, C.; Finkelstein, A.V. The classification and origins of protein folding patterns. Ann. Rev. Biochem. 1990, 57, 1007–1039.
- Kister, A.E.; Finkelstein, A.V.; Gelfand, I.M. Common features in structures and sequences of sandwich-like proteins. Proc. Natl. Acad. Sci. USA 2002, 99, 14137–14141.
- Fraser, R.D.B.; MacRae, T.P.; Parry, D.A.D.; Suzuki, E. The structure of feather keratin. Polymer 1971, 12, 35–56.
- Hallahan, D.L.; Keiper-Hrynko, N.M.; Shang, T.Q.; Ganzke, T.S.; Toni, M.; Dalla Valle, L.; Alibardi, L. Analysis of gene expression in gecko digital adhesive pads indicates significant production of cysteine- and glycine-rich beta keratins. J. Exp. Zool. 2009, 312B, 58–73.
- Calvaresi, M.; Eckhart, L.; Alibardi, L. The molecular organization of the beta-sheet region in corneous beta-proteins (beta-keratins) of sauropsids explains it stability and polymerization into filaments. J. Struct. Biol. 2016, 194, 282–291.
- Fraser, R.D.B.; Parry, D.A.D. Lepidosaur β-keratin chains with four 34-residue repeats: Modelling reveals a potential filament-crosslinking role. J. Struct. Biol. 2020, 209, 107413.
- Holthaus, K.B.; Alibardi, L.; Tschachler, E.; Eckhart, L. Identification of epidermal differentiation genes of the tuatara provides insights into the early evolution of lepidosaurian skin. Sci. Rep. 2020, 20, 12844.
