Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biochemistry & Molecular Biology
Members of the tubulin superfamily are GTPases; the activities of GTPases are necessary for life. The members of the tubulin superfamily are the constituents of the microtubules and the γ-tubulin meshwork. Mutations in members of the tubulin superfamily are involved in developmental brain disorders, and tubulin activities are the target for various chemotherapies. The intricate functions (game) of tubulins depend on the activities of the GTP-binding domain of α-, β-, and γ-tubulin.
- GTPase
- tubulins
- γ-tubulin
- meshwork
- cytoskeleton
1. Introduction
Proteins known as GTPases hydrolyze GTP to realize a variety of functions in both eukaryotes and prokaryotes. GTPases contain a conserved core GTPase domain with two stable states: one that strongly binds to GTP and the second that is either GDP-bound or GTP/GDP-free. The cycling between GTP and GDP is the essence of the GTPases and forms a cellular switch that orchestrates multiple cellular functions [1]. The GDP-to-GTP transition is accompanied by a conformational change that enables GTPases to transduce signals to interacting proteins, transmitting changes to pathways, such as vesicle trafficking or cellular morphology [2]. Therefore, it is not surprising that GTPases’ dysfunction contributes to the development of multiple diseases [3,4].
The activities of GTPases regulate the dynamics of cytoskeletal elements, such as actin, microtubules, and the γ-tubulin meshwork. The Ras homologous (Rho) family of small GTPase (including Rac, Cdc42, and Rho) modify the actin cytoskeleton, whereas heterodimers of the GTPases α- and β-tubulin (α/β-tubulin) are the constituents of microtubules. The binding of GTP to α-tubulin affects the structure of the protein [5,6]; in contrast, the binding and hydrolysis of GTP to β-tubulin regulate the dynamics of microtubules [7]. Another member of the tubulin superfamily, γ-tubulin, is also a GTPase that binds and hydrolyzes GTP [8,9,10]. γ-Tubulin is the major component of the γ-tubulin meshwork [9,10,11,12,13], but the functions of the GTPase activities of γ-tubulin remain elusive. The possible functions of the GTP-binding domain of γ-tubulin as a regulator of the dynamics of the γ-tubulin meshwork are summarized and discussed in this entry based on the latest advances in the field.
2. α-, β-, and γ-Tubulin, and Microtubule Formation
Human γ-tubulin is one of the five known tubulin members (γ-, α-, β-, ε-, and δ-tubulin) in the GTPase superfamily of tubulins [14]. A common feature among the members is a conserved GTP-binding domain in the N-terminal region, which consists of five α-helices and six parallel β-strands (Figure 1) [15,16], whereas the C-terminal region is most variable [3].
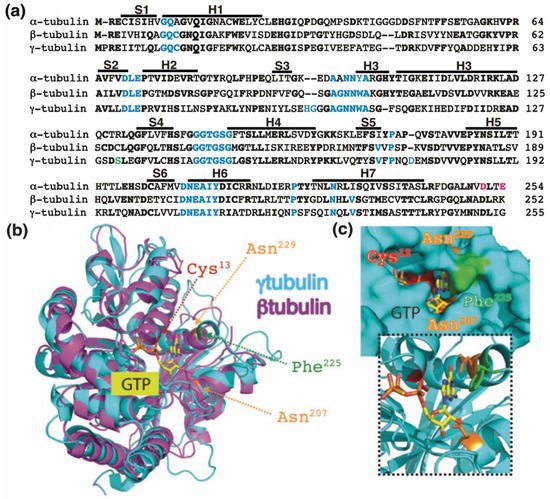
Figure 1. The GTP-binding domain. (a) Sequence alignment of the N-terminal GTP-binding domain (enclosing the first five α-helices and six parallel β-strands [16]) and nearby regulating sequences of human α-tubulin (residues 1–254; GenBank: AAA91576), β-tubulin (residues 1–252; GenBank: AAB59507.1), and γ-tubulin (residues 1–255; GenBank: AAV38734.1). Bold and magenta letters indicate identical and essential residues for the β-tubulin-mediated hydrolysis of GTP, respectively [21]. Residues involved in GTP binding are blue. Ser131 is labeled green. Helices (H) and strands (S) are indicated. (b) The known three-dimensional structures of β-tubulin (magenta) and γ-tubulin (cyan) with a view down the GTP-binding pocket, highlighting Cys13, Asn207, Phe225, and Asn229. (c) The molecular surface of the GTP/GDP-binding pocket of γ-tubulin is displayed, and GTP is shown in a stick representation. The inset (dashed lines) shows GTP together with the protein with only the side-chains of Cys13, Asn207, Phe225 and Asn229 displayed. (b,c) Reproduced with permission from B.O. Villoutreix; published by American Association for Cancer Research, 2015 [30].
3. The GTP-Binding Domain of γ-Tubulin
The homology between the protein sequences of the GTPase domains of α-tubulin (residues 1–254 in Figure 1a) and β-tubulin (residues 1–252 in Figure 1a) is 63%. In contrast, the protein sequence in the GTPase domain of γ-tubulin (residues 1–255 in Figure 1a) exhibits 60%, and 64%, homology with the corresponding sequences in α-tubulin and β-tubulin, respectively, suggesting that the nucleotide-binding site in the GTPase domain of γ-tubulin is most similar to the E site (β-tubulin) rather than to the N site (α-tubulin).
The crystal structure of γ-tubulin is similar to the structures of α- and β-tubulin (Figure 1b) [16,25,31]. Compared with the N terminal region of α- and β-tubulin, γ-tubulin has one deletion and three insertions (Figure 1a). γ-Tubulin lacks a His residue between residues 107 and 108; this probably alters the α-helix structure formed by the amino acid sequence in this region [25]. The insertions of His-Gly and Asp residues at position 96 and 97 and at position 176, respectively, are part of disorder regions (with not define secondary structure) in the γ-tubulin structure and might be involved in longitudinal contact of the surface of γ-tubulin with other proteins [25].
4. The Dynamics of the γ-Tubulin Meshwork
There are many characteristics that are shared among the members of the tubulin superfamily; together, the members are important proteins in the development of the brain as well as in the fight against cancer [47,48,49,50,51,52]. The structural plasticity and dynamics of microtubules depend on the activities of the GTP-binding domain of α-, β-, and γ-tubulin [8,53]. Similarly, one should expect that the GTP-binding domain in tubulins also regulates the structural plasticity and dynamics of the γ-tubulin meshwork.
Both γ-tubulin and its GTP-binding domain are necessary for cell survival [8,9,10]. Though, it is unclear how the GTPase contributes to the function of the protein. The canonical functions of γ-tubulin start in the S phase with the duplication of centrosomes that, at the onset of mitosis, ensures the assembly of a bipolar mitotic spindle and the segregation of sister chromatids between offspring cells. γTURCs also nucleate cytosolic microtubules in the interphase. Apart from these well-studied functions, γ-tubulin forms a meshwork. However, we know too little about how the meshwork is regulated, its functions, and its dynamics. The problems in elucidating the functions of γ-tubulin are that many of the cell structures containing γ-tubulin are not preserved after preparation of the samples [35], and available antibodies do not recognize the whole cellular pool of γ-tubulin [12,42]. In addition, a cell model with a total CRISPR-Cas9-mediated knockout of γ-tubulin is impossible to achieve as γ-tubulin is essential for cell survival [10]. In contrast, short hairpin (sh)RNA-mediated reduction of γ-tubulin expression results in stable cell lines that express half the amount of the total γ-tubulin pool [36], providing a useful tool for studying the functions of the γ-tubulin meshwork [10,36].
Thus, to increase our knowledge of the meshwork, live imaging of cells with a fluorescence label meshwork may provide us with hints on the biological functions of γ-tubulin and its GTPase. In time-lapse images of living U2OS cells that stably express both TUBG-shRNA (which lowers the endogenous pool of γ-tubulin by ~50% [36]) and a C-tagged TUBG1-green fluorescence protein (GFP) shRNA-resistant gene (which fluorescence labels the γ-tubulin meshwork [10,43]), it is visible that, during the interphase, γ-strings are found in the cytoplasm and nucleus, whereas centrosomes and γ-tubules are in the cytosol and their positions are constantly changing (Figure 3 and Video S1) [11,43]. It is apparent that γ-tubules can be formed close to the nuclear envelope and that centrosomes can either nucleate γ-tubules or move along and among them (Figure 3 and Videos S1 and S2). The location of the centrosomes varies from cell to cell. Centrosomes are located on the cytosolic side of the nuclear envelope or in the cytosol, but in both cases, their position is constantly changing (Figure 3 and Videos S1 and S2). The number, location, and length of γ-tubules also vary in both cells and overtime (Figure 3 and Videos S1 and S2).
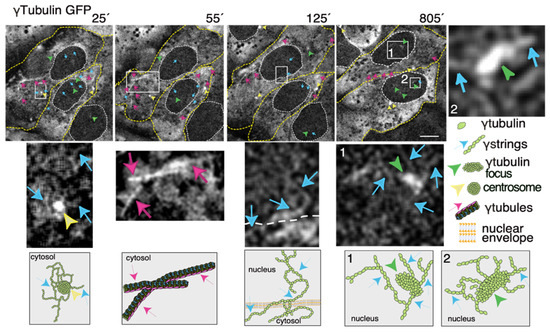
Figure 3. The dynamics of the γ-tubulin meshwork in living cells. Time-lapse series of fluorescence images showing U2OS cells stably coexpressing TUBG-short hairpin (sh)RNA and GFP-tagged sh-resistant γ-tubulin (γtubulin GFP). Images present chosen frames illustrating the changes in the position of γ-strings, γ-tubules, centrosomes, and γ-tubulin foci during the interphase. The outer membrane of the cells and the nuclei are indicated by dotted yellow and white lines, respectively. Yellow arrowheads, blue arrows, and magenta arrows indicate centrosomes, γstrings, and γ-tubules, respectively. Green arrowheads denote nucleating γ-tubulin foci. The images shown were collected every 5 min and represent selected frames showing the changes that γstrings, γ-tubules, centrosomes, and γ-tubulin foci undergo during interphase. White boxes indicate the areas magnified in the inset. Scale bars: 10 μm. The lower panels are schematic representations of the structures shown in the corresponding white box. See Videos S1–S3. The stable cell line was obtained, and time-lapse experiments were performed as previously described [10,12,35]. These data are from unpublished works by Alvarado-Kristensson et al.
γ-Strings span from the cytosolic compartment through the nuclear membrane into the chromatin and can emanate from centrosomes and γ-tubulin rich structures (Figure 3 and Videos S1 and S3). These foci localize to both the cytoplasm and nucleus and γ-strings can emanate from those structures (Figure 3 and Videos S1 and S3). Notably, the nuclear foci are located towards the interior, where genes normally are positioned (Figure 3 and Videos S1 and S3) [32]. An earlier study using various mammalian cell lines demonstrated that γ-tubules nucleate on cytosolic aggregates or γTURC foci together with pericentrin [9]. Both γTURCs and pericentrin are cytosolic proteins, which strongly suggests that the contents of the nuclear and cytosolic foci differ, but the common component is γ-tubulin [9].
This entry is adapted from the peer-reviewed paper 10.3390/cells10040745
This entry is offline, you can click here to edit this entry!
