The main sunflower breeding goals are aimed towards high seed and oil yield, genetic resistance and high level of tolerance to the economically most important diseases, insects and parasitic weed (broomrape), as well as tolerance to abiotic stresses (in the first place to drought). As one of the most important oilseed crops worldwide, in order to meet growing global demands for sunflower products, intensified efforts for implementation of all available advanced breeding tools are required to improve the quantity and the quality of sunflower output by focusing on factors that are limiting phenotype expression of genetic potential. Special attention should be paid to the complexity in inheritance of the afore-mentioned traits, especially for resistance and tolerance to different pests and drought.
- sunflower
- genomics
- epigenomics
- integrated omics
1. Introduction
Crop production today is threatened by severe abiotic stresses due to extreme weather conditions (droughts, floods and other disasters), accompanied by emerging diseases and a decrease in arable land [1][2][3]. Certainly, the most important mission in agriculture is to provide sufficient quantities of plant based products for a growing world population. The benefits in yield and food quality brought by “The Green Revolution” are far from enough to keep up with the pace as the increasingly growing demand forecasts an increase of 70% for food requirements by 2050 [4]. Projection of linear progress of 2% of genetic gain in order to meet demands is questionable as, so far, annual gain in crop productivity is rated from 0.8 to 1.2%, which is considered insufficient [5].
Native to North America, with its exceptional ability for adaptability to different climatic and soil conditions, sunflower is grown around the globe as a crop that significantly contributes in vegetable oil consumption. The main sunflower breeding goals are aimed towards high seed and oil yield, genetic resistance and high level of tolerance to the economically most important diseases, insects and parasitic weed (broomrape), as well as tolerance to abiotic stresses (in the first place to drought). As one of the most important oilseed crops worldwide, in order to meet growing global demands for sunflower products, intensified efforts for implementation of all available advanced breeding tools are required to improve the quantity and the quality of sunflower output by focusing on factors that are limiting phenotype expression of genetic potential. Thus far, substantial progress has been made in order to improve breeding process in sunflower by application of DNA markers, especially for disease resistance [6] and tolerance to abiotic stress [7].
2. Molecular Omics Profiling
In this regard, one of the drawbacks is the insufficient information on existing genetic resources, which has the consequence that although there is a very large collection of genetic material around the world, there is a lack in discovering beneficial alleles that can be utilized in breeding and transferred into elite genotypes [3][8]. As previously outlined, detection of favorable genetic variation largely depends on thorough genome sequencing through broad and deep resequencing and construction of pangenome, in order to characterize in detail diverse germplasm, thus providing clear profile view of the locked genetic variation [9]. Since the development of the first genetic map on wild sunflower in 1993, the evolution of molecular markers enabled the successive addition of new markers to the map and enabled the positioning and detection of desirable genes on individual linking groups [10][11][12][13]. Finally, a high-quality reference for the sunflower genome is available (Table 1) that contains 3.6 gigabases consisting of long and highly similar repeats and allows more efficient exploitation of sunflower genetic background towards improvement in biotic and abiotic stress resistance, as well as oil production [14].
Table 1. Sunflower genome and pangenome main characteristics.
| Composition Type | Accessions | Strategy | Size | Reference |
|---|---|---|---|---|
| Genome | Inbred line XRQ | 102× sequencing coverage of the genome of the inbred line XRQ using 407 single-molecule real-time (SMRT) cells on the PacBio RS II platform. | 52,232 protein-coding genes 5803 spliced long non-coding RNAs |
[14] |
| Pangenome | 493 sunflower accessions which include: 287 cultivated lines, 17 Native American landraces and 189 wild accessions representing 11 compatibile wild species | Pangenome assembled through de novo assembly of unmapped reads |
61,205 genes | [15] |
As indicated in the previous study, an assembly of three high-quality sunflower reference genomes is available, two of them covering genomes of inbred lines XRQ and HA412-HO and one of the restorer line PSC8 These high resolution sunflower maps enable narrowing the targeted area in the pursuit for desirable genes for many important traits [16]. In order to obtain more complete information on the total genetic variability of a particular species, the concept of pangenome has recently been disguised (Figure 1). It is based on the fact that the total genetic variation of a population or species consists of a core genome that represents a set of genes that are common to all individuals and a dispensable genome consisting of a small number of genes that are absent in one or more individuals [17][18].
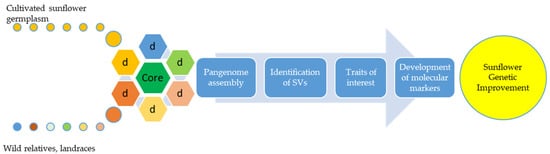
Figure 1. The basic concept of pangenome applied to sunflower. The concept of pangenome can be used for broadening genetic diversity in the pursuit for important traits. Using biotechnological tools, molecular markers can be developed for the appropriate trait and used in breeding to improve sunflower genetics.
Cultivated sunflower is related to a large number of wild sunflower species. By applying technological progress, it is possible to overcome difficulties in the use of wild species [19]. As a useful example, CRISPR–Cas9 genome editing strategy was applied for editing several loci important for yield and productivity in cultivated tomato lines and enabled de novo domestication of wild tomato [20]. By comparison of assembled genes with wild relatives, they were able to identify introgressed genomic regions from wild sunflower species.
These reversible modifications of the genomic DNA have significant functions in gene management and cell activities [21]. Epigenetic studies have gained wings in recent years through high-throughput assays and provided evidence of the role of epigenetic DNA marks on phenotypic expression of several traits [3]. Although several epigenetic marks (known as tags) have been discovered, the mainly characterized ones are DNA methylation and histone modification [21]. Furthermore, epigenetic variations in DNA methylation are inducing epiallelic diversity, which is responsible for phenotypic variation via changes in transcription and morphology.
3. Integrated Omics Approach—Systems Biology
Technological developments enabled detailed and complex omic-studies and provided valuable inferences generated by examining the functioning of the cellular system under different circumstances. Such data can be utilized to formulate predictive models of behavior of important agronomic traits and integrate them within the concept of quantitative genetics [3]. As outlined, omics data are expected to group interaction information within and between different biological layers and enhance the predictive ability of a particular trait [22]. An integrative approach, which includes data from different omics datasets, is known as systems biology [23][24].
One of the flaws of this system is a confusion with the correct interpretation of a huge amount of omics data, often without a clear connection [25]. To overcome incorrect interpretation of data, systematic multi-omics integration (MOI) with a well-defined scheme for linking different data is proposed. An excellent example of the application of the integrated approach is given in a comprehensive study where different predictors from genomic, transcriptomic and metabolomic data, measured on maize parent lines, were applied in order to predict the effects of untested hybrids for important quantitative traits [22]. As an outcome, combining omics with genomic data improved predictive ability, while in comparison between predictors, transcriptomic data outperformed others.
4. Conclusions and Prospects
In this regard, a detailed characterization of genotypic and phenotypic diversity is very important in order to have a clearer insight into the variations of important traits, which can be utilized to improve sunflower genetics. Lately, intensive research to improve prediction accuracy has resulted in the extensive use of different machine learning techniques. Being based on how humans learn and process information, machine learning is a powerful tool for processing complex data for accurate prediction and have already been used for precision breeding [26]. Considering the future advancement of sunflower as one of the most important oil crops worldwide, integrated model implementation should also include sunflower researcher community from different institutions and development of effective teamwork with the aim of doing a comprehensive integration of available technologies and data information as the best way to provide beneficial prospect.
This entry is adapted from the peer-reviewed paper 10.3390/plants10061150
References
- Sedeek, K.E.M.; Mahas, A.; Mahfouz, M. Plant Genome Engineering for Targeted Improvement of Crop Traits. Front. Plant Sci. 2019, 10, 114.
- Varshney, R.K.; Sinha, P.; Singh, V.K.; Kumar, A.; Zhang, Q.; Bennetzen, J.L. 5Gs for crop genetic improvement. Curr. Opin. Plant Biol. 2020, 56, 190–196.
- Scossa, F.; Alseekh, S.; Fernie, A.R. Integrating multi-omics data for crop improvement. J. Plant Physiol. 2021, 257, 153352.
- Fischer, T.; Byerlee, D.; Edmeades, G. Crop Yields and Global Food Security: Will Yield Increase Continue to Feed the World; ACIAR Monograph: Canberra, Australia, 2014; p. 634.
- Li, H.; Rasheed, A.; Hickey, L.T.; He, Z. Fast-Forwarding Genetic Gain. Trends Plant Sci. 2018, 23, 184–186.
- Imerovski, I.; Dedić, B.; Cvejić, S.; Miladinović, D.; Jocić, S.; Owens, G.L.; Kočiš Tubić, N.; Rieseberg, L.H. BSA-seq mapping reveals major QTL for broomrape resistance in four sunflower lines. Mol. Breed. 2019, 39, 41.
- Balliau, T.; Durufle, H.; Blanchet, N.; Blein-Nicolas, M.; Langlade, N.B.; Zivy, M. Proteomic data from leaves of twenty-four sunflower genotypes under water deficit. OCL 2021, 28, 12.
- Mascher, M.; Schreiber, M.; Scholz, U.; Graner, A.; Reif, J.C.; Stein, N. Genebank genomics bridges the gap between the conservation of crop diversity and plant breeding. Nat. Genet. 2019, 51, 7, 1076–1081.
- Xu, Y.; Li, P.; Zou, C.; Lu, Y.; Xie, C.; Zhang, X.; Prasanna, B.M.; Olsen, M.S. Enhancing genetic gain in the era of molecular breeding. J. Exp. Bot. 2017, 68, 2641–2666.
- Rieseberg, L.H.; Choi, H.; Chan, R.; Spore, C. Genomic map of a diploid hybrid species. Heredity 1993, 70, 285.
- Gedil, M.A.; Wye, C.; Berry, S.T.; Seger, B.; Peleman, J.; Jones, R.; Leon, A.; Slabauh, M.B.; Knapp, S.J. An integrated restriction fragment lenght polimorphism-amplified fragment length polimorphism linkage map for cultivated sunflower. Genome 2001, 44, 213–221.
- Yu, J.K.; Tang, S.; Slabaugh, M.B.; Heesacker, A.; Cole, G.; Herring, M.; Soper, J.; Han, F.; Chu, W.-C.; Webb, D.M.; et al. Towards a saturated molecular genetic linkage map for sunflower. Crop Sci. 2003, 43, 367–387.
- Tang, S.; Kishore, V.K.; Knapp, S.J. PCR-multiplexes for a genome-wide framework of simple sequence repeat marker loci in cultivated sunflower. Theor. Appl. Genet. 2003, 107, 6–19.
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Briere, C.; Owens, G.L.; Carrere, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152.
- Hubner, S.; Bercovich, N.; Todesco, M.; Mandel, J.R.; Odenheimer, J.; Ziegler, E.; Lee, J.S.; Baute, G.J.; Owens, G.L.; Grassa, C.J.; et al. Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat. Plants 2019, 5, 54–62.
- Dimitrijević, A.; Horn, R. Sunflower Hybrid Breeding: From Markers to Genomic Selection. Front. Plant Sci. 2018, 8, 2238.
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955.
- Khan, A.W.; Garg, V.; Roorkiwal, M.; Golicz, A.A.; Edwards, D.; Varshney, R.K. Super-Pangenome by Integrating the Wild Side of a Species for Accelerated Crop Improvement. Trends Plant Sci. 2020, 25, 148–158.
- Warschefsky, E. Back to the wilds: Tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. Am. J. Bot. 2014, 101, 1791–1800.
- Zsogon, A.; Čermak, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216.
- Großkinsky, D.K.; Syaifullah, S.J.; Roitsch, T. Integration of multi-omics techniques and physiological phenotyping within a holistic phenomics approach to study senescence in model and crop plants. J. Exp. Bot. 2018, 69, 825–844.
- Westhues, M.; Schrag, T.A.; Heuer, C.; Thaller, G.; Utz, H.F.; Schipprack, W.; Thiemann, A.; Seifert, F.; Ehret, A.; Schlereth, A.; et al. Omics-based hybrid prediction in maize. Theor. Appl. Genet. 2017, 130, 1927–1939.
- Budak, H.; Hussain, B.; Khan, Z.; Ozturk, N.Z.; Ullah, N. From genetics to functional genomics: Improvement in drought signaling and tolerance in wheat. Front. Plant Sci. 2015, 6, 1012.
- Wu, S.; Ning, F.; Zhang, Q.; Wu, X.; Wang, W. Enhancing Omics Research of Crop Responses to Drought under Field Conditions. Front. Plant Sci. 2017, 8, 174.
- Jamil, I.N.; Remali, J.; Azizan, K.A.; Nor Muhammad, N.A.; Arita, M.; Goh, H.-H.; Aizat, W.M. Systematic Multi-Omics Integration (MOI) Approach in Plant Systems Biology. Front. Plant Sci. 2020, 11, 944.
- Weckwerth, W.; Ghatak, A.; Bellaire, A.; Chaturvedi, P.; Varshney, R.K. PANOMICS meets germplasm. Plant Biotechnol. J. 2020, 18, 1507–1525.
