Melanogenesis is the process leading to the synthesis of melanin, the main substance that influences skin color and plays a pivotal role against UV damage. Altered melanogenesis is observed in several pigmentation disorders. Melanogenesis occurs in specialized cells called melanocytes, physically and functionally related by means of autocrine and paracrine interplay to other skin cell types. Several external and internal factors control melanin biosynthesis and operate through different intracellular signaling pathways, which finally leads to the regulation of microphthalmia-associated transcription factor (MITF), the key transcription factor involved in melanogenesis and the expression of the main melanogenic enzymes, including TYR, TYRP-1, and TYRP-2. Epigenetic factors, including microRNAs (miRNAs), are involved in melanogenesis regulation. miRNAs are small, single-stranded, non-coding RNAs, of approximately 22 nucleotides in length, which control cell behavior by regulating gene expression, mainly by binding the 3′ untranslated region (3′-UTR) of target mRNAs.
- melanocyte
- melanogenesis
- microRNA
- skin pigmentation
1. Introduction
Skin represents the primary line of defense against environmental stressors, including chemical stimuli, microbial insults, allergens, and ultraviolet (UV) radiation. Protection from UV rays is essentially based on melanogenesis, the process leading to the synthesis of pigments called melanin, the main substance that influences skin color. Melanin protects the skin from harmful UV rays, as it can absorb UV and visible light and shows antioxidative and radical scavenging abilities, limiting UV-induced effects on cellular macromolecules, mainly DNA, thus protecting cells from genotoxic damage [1]. Therefore, reduced melanogenesis is also a major risk factor for melanoma and other skin cancers [2][3]. Nevertheless, increased melanogenesis and melanin accumulation is associated with hyperpigmentation disorders [4][5]. Skin hyperpigmentation, often associated with aging, hormonal changes, and UVB, is very common in clinical dermatology and includes dermal conditions such as melasma, chloasma, freckles, age spots, and sunspots [5]. In addition, both hyperpigmentation and hypopigmentation, as observed in vitiligo lesions, are frequently the consequence of inflammation, induced by skin stressors [6]. Indeed, the modulation of skin pigmentation still represents a challenge in treating dermatological disorders, despite several studies having investigated potential cures [7][8]. Melanin is produced by highly specialized cells called melanocytes that are in strict contact with other skin cells, especially keratinocytes. The process of skin coloration consists of melanin biosynthesis and the translocation of melanosomes, small organelles containing melanin, from melanocytes into epidermal keratinocytes [6][7][9].
Melanogenesis is a very complex process which includes the development, survival, and differentiation of melanocytes. It involves more than 150 genes and several signaling pathways that operate both at transcriptional and post-transcriptional levels in regulating the main melanogenic molecular players. These include transcription factors, enzymes, and regulatory molecules produced by melanocytes, as well as other skin cells including keratinocytes, dermal fibroblasts, and inflammatory and endocrine cells [6][7][10][11].
Altered gene expression and melanogenic regulatory factor activities are involved in melanogenesis dysfunction [4][11]. Increasingly, studies indicate that gene expression is also influenced by several epigenetic events, including chromatin modification, DNA methylation, and non-coding RNA classes such as long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) [12][13][14][15]. Indeed, the role of miRNAs in melanogenesis has been widely investigated.
2. Melanin Biosynthesis
Melanin synthesis occurs in highly specialized cells called melanocytes, localized in the basal layer of the epidermis and hair follicles [16][17]. Melanocytes consist of several ramifications called dendrites that end in keratinocytes, and each melanocyte is strictly connected to more than 30 keratinocytes, constituting the melanogenic unit [18]. During melanogenesis, a series of sequential reactions synthesize melanin, which is translocated into neighboring keratinocytes by means of melanosomes [6][7][9].
Melanin production is controlled by several enzymes including tyrosinase (TYR), tyrosine hydroxylase I (THI), and phenylalanine hydroxylase (PAH) in the initiation phase of melanin synthesis. Tyrosinase-associated protein 1 (TYRP-1) and tyrosinase-associated protein 2 (TYRP-2), also called dopachrome tautomerase (DCT), operate in the later phase [4][19]. TYR is a membrane-bound glycoprotein which plays a key role in the process, as it is considered the rate-limiting enzyme for melanin biosynthesis (Figure 1).
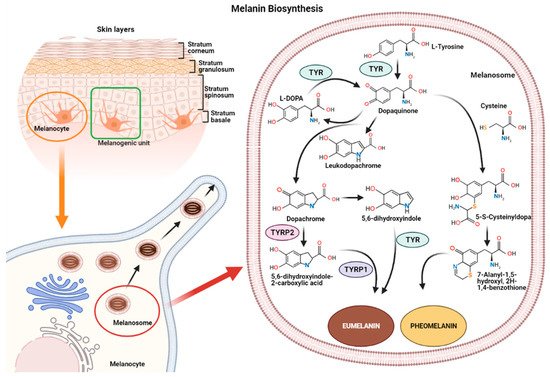
Figure 1. Representation of the melanogenic unit and melanin synthesis in melanosomes (left). Schematic representation of eumelanin and pheomelanin biosynthetic pathways (right).
The reaction catalyzed by TYR leads to L-tyrosine being transformed into dopaquinone by oxidation [20]. Dopaquinone is highly reactive and can follow two reaction chains from which eumelanin and pheomelanin originate:
-
(1) In reactions leading to eumelanin production, dopaquinone undergoes intramolecular cyclization to produce leukodopachrome (cyclodopa). Cyclodopa undergoes redox exchange with another molecule of dopaquinone to form dopachrome and DOPA [20]. The dopachrome downstream process is branched in two ways. The first leads to the formation of 5,6-dihydroxyindole-2-carboxylic acid (DHICA) through TYRP-2 intervention and then into eumelanin by TYRP-1 conversion. The second leads to the conversion of dopachrome into 5,6-dihydroxyindole (DHI) and then into eumelanin involving TYR. At the end of this reaction, black-brownish eumelanin is formed.
-
(2) In reactions leading to pheomelanin production, in the presence of cystein or glutathione, dopaquinone can be converted into 5-S-cysteinyldopa, or glutathionyldopa, which is then converted into quinoline and finally polymerized into red-yellow pheomelanin.
In the skin, both eumelanin and pheomelanin form complex heteropolymers. The total amount of melanin, as well as the ratio between eumelanin to pheomelanin, are considered to determine skin color. Indeed, the role of the ratio between eumelanin and pheomelanin is still under debate, as many authors indicate that it remains unchanged in various phototypes of human skin and, thus, skin color is dependent on total melanin amount [4][21]. Synthesized melanin is collected into melanosomes, which follow a complex maturation process and are transported to keratinocytes along actin filaments in association with motor proteins [4][22].
3. Melanogenesis Regulation
Several stimuli control melanogenesis, including external factors such as UV rays or environmental pollution [11]. Furthermore, a series of endogenous molecules released by melanocytes, keratinocytes, dermal fibroblasts, and immune cells modulate the process paracrinally and autocrinally. These are mainly the α-melanocyte stimulating hormone (α-MSH) and the adrenocorticotropic hormone (ACTH), derived from the cleavage of pro-opiomelanocortin (POMC), from both melanocytes and keratinocytes [11][23]. In addition, stem cell factor (SCF), peptide endothelin 1 (ET-1), hepatocyte growth factor (HGF), keratinocyte growth factor (KGF), basic fibroblast growth factor (bFGF), and inflammatory mediators such as cytokines, prostaglandin E2 (PGE2), and nitric oxide (NO) significantly regulate melanogenesis [24][25]. All these stimuli operate with the activation of several signaling pathways [9][26]. Detailed descriptions of the multiple pathways in melanogenesis have been recently reported by others [6][7][9][11]. Here, the main signaling pathways are briefly summarized (Figure 2).
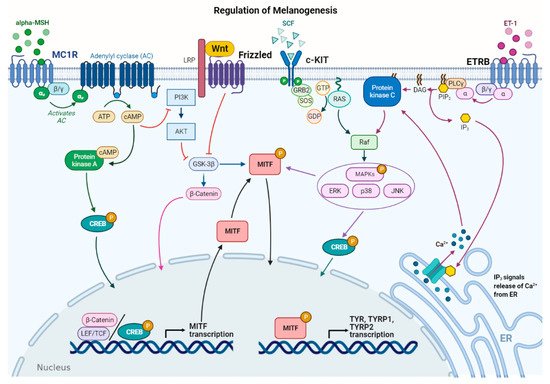
Figure 2. Main signaling pathways involved in melanogenesis regulation. α-melanocyte stimulating hormone (α-MSH) binds to melanocortin 1 receptor (MC1R), which increases cAMP levels, activating PKA and PI3K/AKT pathway. The former phosphorylates CREB protein, promoting MITF transcription; the latter interplays with Wnt/β-catenin pathway by phosphorylating GSK-3β, which, in turn, releases β-catenin to promote MITF transcription. Stem cell factor (SCF) binds to c-KIT receptor, activating MAPK pathway and phosphorylating CREB. Peptide endothelin 1 (ET-1) binds to its receptor, ETRB, activating PKC and stimulating MITF transcription. MITF is phosphorylated at the post-transcriptional level to promote transcription of the melanogenic enzymes.
In melanocytes, signaling pathways regulating melanogenesis operate through membrane receptors with different molecular activities. These receptors include G protein-coupled receptors (GPCRs) such as the melanocortin-1 receptor (MC1R), which is mainly expressed in melanocytes [27], adrenergic receptors, endothelin type B receptor (ETRB), frizzled receptor (FZD), and tyrosine kinase receptors such as tyrosine kinase receptor KIT, bFGF, and HGF receptors [7][28]. Most signaling pathways lead to the regulation of microphthalmia-associated transcription factor (MITF), a basic helix–loop–helix leucine zipper (bHLH-ZIP) transcription factor. MITF is the dominant transcription factor in melanogenesis as it controls melanocyte development, survival, and proliferation, as well as the steps involved in melanin synthesis [29][30]. MITF induces the transcription of the melanogenic genes, including TYR, TYRP-1, and TYRP-2, by binding to the conserved consensus elements of the promoter regions [30]. Moreover, MITF regulates several other genes involved in melanogenesis, including those required to control melanosome maturation, traffic, and distribution to keratinocytes [31]. Due to the prominent role of MITF, regulation of its expression and activity represents a key event in melanogenesis. At the transcriptional level, MITF expression is regulated by several transcriptional factors that bind the MITF promoter, including cyclic adenosine monophosphate (cAMP)-response element binding protein (CREB), paired box family of transcription factor 3 (PAX3), sex determining region Y-box 9 and 10 (Sox9, Sox10), and Wnt/β-catenin pathway effector lymphoid enhancer-binding factor 1 (LEF-1) [7][30].
Furthermore, MITF activity is regulated at the post-transcriptional level mainly by phosphorylation [32]. MITF may be phosphorylated by several kinases such as MAPK, p38, ribosomal S6 kinase (RSK), and GSK3β [7][11][33][34]. MITF phosphorylation can favor the recruitment of transcriptional coactivator CBP/P300 of CREB, thus increasing the transcription of TYR, TYRP-1, and TYRP-2 melanogenic enzymes [30]. The MAPK signaling pathway can be activated by several extracellular factors, which operate through tyrosine kinase receptors including SCF, a relevant signal produced by keratinocytes and fibroblasts, and HGF and bFGF, mainly produced by keratinocytes [35]. Notably, melanogenesis-related signaling pathways may be positively or negatively modulated by several inflammatory factors that finally regulate skin pigmentation. This regulation is very complex as some molecules have a stimulatory effect on melanogenesis, while others show inhibitory effects. In addition, the production of these inflammatory mediators depends on the communication between several epidermal and dermal cells, as well as specific stimuli inducing inflammation [6].
As mentioned above, UV rays represent external stimuli with significant effects on melanogenic signaling pathways, resulting in increased melanin production. Several cellular and molecular processes are involved in the UV–skin reaction. Indeed, UV irradiation induces MITF expression and consequently melanogenic gene expression [3][30]. Acquired knowledge shows that UV irradiation influences different skin cell types, including melanocytes, keratinocytes, and dermal fibroblasts. UV-induced melanogenesis has been associated with the release of several molecules, including α-MSH, ACTH, SCF, and ET-1, which regulate the signaling pathways described above [24][35][36][37].
4. miRNA Activities and Identification
MicroRNAs (miRNAs) represent a class of non-coding RNAs of approximately 21–23 nucleotides in length. Since their discovery in 1993 [38], miRNAs have been largely studied as essential regulators of gene expression. miRNAs are highly conserved among species and are found in different cell types and organisms, including plants, animals, and viruses [39][40]. It is now estimated that miRNAs target approximately 60% of genes in humans and other mammals [41]. Due to their role in mRNA expression regulation, miRNAs are involved in all cellular activities, including proliferation, differentiation, migration, apoptosis, and immune responses, both in normal and pathological conditions [42][43][44][45]. Several mechanisms ensure high stability in miRNAs. Since they can be easily detected in almost all bodily fluids, miRNAs are now considered important biological markers and potential therapeutic molecules [46]. Inside cells, miRNAs derive from long precursor transcripts which give rise to mature miRNAs through a multi-step process and are then incorporated into an RNA–protein complex known as the RNA-Induced Silencing Complex (RISC) [47]. miRNA activity mainly induces gene expression reduction by binding to sequences in the 3′ untranslated region (3′-UTR) of target genes. This can lead to mRNA target degradation, or inhibition of translation and reduction in protein levels [44]. However, miRNA activities show higher complexity as each miRNA can target different genes and several miRNAs can regulate the same gene’s expression. Moreover, the strict interplay between long non-coding RNAs (lncRNAs) and miRNAs contributes to gene expression regulation [39][44][48].
In general, from a methodological point of view, investigations into miRNA functions and roles in a selected process include a preliminary phase of miRNA identification using high-throughput methods such as microarray, followed by target gene identification with a bioinformatics approach. Finally, target genes need to be validated by 3′-UTR reporter assays and analysis conducted on induced cellular effects following transfection experiments with miRNA mimics and antagomiRs [49]. miRNAs are involved in skin development and functions both in normal and pathological conditions [50]. Furthermore, increasing evidence shows that selected miRNAs regulate specific events occurring in the skin, such as melanogenesis, as detailed below [51][52].
4.1. miRNA Regulating MITF
Due to the key role of MITF as an essential regulator of the melanogenic process, several studies have investigated miRNAs with potential effects on MITF expression and consequent regulation of mRNA levels in melanogenic enzymes [3][29][30][53] (Table 1). Some preliminary studies concerning melanogenesis regulation by miRNAs have been performed in fiber-producing animals, such as alpaca, due to the contribution of melanin synthesis to coat color and the specific interest of alpaca breeders in animal color coat modulation. Differences in miRNA profiles from alpaca skins with different colored coats were identified and most differentially expressed miRNAs showed predicted targets involved in pigmentation [54][55]. These results led to further investigation concerning the functional role of selected miRNAs. Data obtained by Zhu Z et al. (2010) demonstrated the functional role of miR-25 in reducing MITF mRNA and protein, TYR, and TYRP-1 expression in cultured melanocytes. In addition, an inverse relationship was observed between miR-25 level and coat color. Similarly, in alpaca melanocytes, miR508-3p also directly targets MITF, binding to the 3′-UTR of the gene. miR508-3p overexpression downregulated MITF expression, resulting in a decrease in TYR, TYRP-2, and melanin production [56]. Interestingly, in 2012, Dong C. et al. investigated the role of miR-137, another miR-targeting MITF, in a transgenic mice model [57]. Initially investigating melanoma cells lines, where its overexpression downregulated MITF [58], it was found that miR-137 also decreased the expression of the MITF protein and its downstream genes in transgenic mice. Notably, miR-137 had an impact on coat color, demonstrating that modulating a specific miR may significantly regulate melanogenesis, at least in animal models [57].
Table 1. microRNAs regulating MITF in melanogenesis.
| miRNA | Cell Model | Target Gene | Effect on Melanogenesis |
Ref. |
|---|---|---|---|---|
| miR-25 | Alpaca melanocytes | MITF | Negative | [56] |
| miR508-3p | Alpaca melanocytes | MITF | Negative | [56] |
| miR-137 | Alpaca melanocytes | MITF | Negative | [58] |
| miR-675 | Melanocytes of melasma patients, keratinocytes of melasma | MITF | Negative | [5][59][60] |
| miR-218 | Melan-a murine melanocytes, human skin OTC | MITF | Negative | [61] |
| miR-183 | B16 melanoma cells | MITF | Negative | [62] |
| miR-340 | Human epidermal melanocytes (Pig-I) | MITF | Negative | [63][64] |
| miR-200a-3p | B16-4A5 melanoma cells | MITF | Negative | [52] |
| miR-148a-3p | B16-4A5 melanoma cells | MITF | Negative | [52] |
| miR-141-3p | B16-4A5 melanoma cells | MITF | Negative | [52] |
Other studies have identified miRNAs regulating skin pigmentation in human melanocytes. Particular interest has been shown in miR-675, another miR which targets MITF and was to be found expressed at low levels in the hyperpigmented skin of melasma patients. In melanocytes or keratinocytes derived from melasma patients, miR-675 upregulation decreased TYRP-1 and TYRP-2 expression, whereas its knockdown increased their expression. Interestingly, miR-675 was also identified in exosomes released from keratinocytes into the extracellular environment [5][59][60]. Another miR involved in melanogenesis by binding the 3′-UTR in MITF is miR-218, which downregulated TYR, TYRP-1, and DCT mRNA and protein levels, reducing melanin content in immortalized melan-a murine melanocytes. In agreement with these data, miR-218 also suppressed melanogenesis in human pigmented skin organotypic culture (OTC) through MITF repression. Anti-miR-218 was also found to promote melanogenesis in human primary melanocytes [61].
Recently, other miRNAs, including miRNA-183 cluster, miR-340, miR-141-3p, and miR-200a-3p, have been investigated for their role in regulating MITF expression and melanogenesis [62]. Results of this research show that the miRNA-183 cluster targets the 3′-UTR of MITF in B16 mouse melanoma cells. miRNA-183 cluster overexpression decreased MITF, TYR, TYRP-1, and DCT expression and melanin production. Conversely, knockdown of the miRNA-183 cluster increased MITF, TYR, TYRP-1, and DCT expression and, consequently, melanin levels. The miR-183 cluster was also involved in regulating the MEK/ERK signaling pathway implicated in cell proliferation and migration, by modulating mitogen-activated protein kinase 1 (MEK1), extracellular regulated protein kinases1/2 (ERK1/2), and CREB expression [62]. miR-340, which was first identified in melanoma cell lines for its ability to bind specifically to the 3′-UTR of MITF [64], has been also investigated in immortalized human epidermal melanocytes (Pig-1), where it downregulates MITF expression and melanin synthesis [63]. Recently, miR-141-3p and miR-200a-3p have been identified as MITF regulators [52]. In this study, comparing miRNA expression profiles in B16-4A5 mouse melanoma cells which were treated or untreated with α-MSH led to 13 miRNAs being identified as differentially expressed through miRNA array analysis. miR-141-3p, miR-200a-3p, and miR-148a-3p, which target MITF, were downregulated in α-MSH-stimulated cells when compared to untreated cells. Furthermore, miR-141-3p and miR-200a-3p overexpression suppressed MITF expression and TYR activity in B16-4A5 cells. Notably, the inhibitory effect on melanin synthesis was confirmed in a three-dimensional tissue culture model of the human epidermis (3D-MHE model) [52].
4.2. miRNAs Regulating Other Genes in Melanogenesis
Besides the abovementioned studied group of miRNAs, with significant regulatory roles in MITF, other miRNAs are involved in melanogenesis by regulating the expression of other molecular targets, including melanogenic enzymes, transcription factors, or components of the signaling pathways which regulate melanogenesis (Table 2).
Several miRNAs, including miR-450b-5p, miR-1208, miR-326, miR-434-5p, miR330-5p, miR-125, miR-145, and miR-203, have been predicted as targeting TYR and most also target other genes, including MITF [65][66].
Table 2. Other microRNAs involved in melanogenesis.
| miRNA | Cell Model | Target Gene | Effect on Melanogenesis |
Ref. |
|---|---|---|---|---|
| miR-434-5p | Mouse skin, human skin cell cultures | TYR | Negative | [65] |
| miR-330-5p | Melanoma cells, normal human melanocytes | TYR | Negative | [56][67] |
| miR-203 | Keratinocytes exposed to UV | Kinesin Superfamily Protein 5b | Positive | [67] |
| miR-3196 | Keratinocytes exposed to UV | Unknown target gene | Positive | [67] |
| miR-21a-5p | Human melanocytes | SOX5 | Positive | [68] |
| miR-145 | Murine melan-a melanocytes | Myo5a | Negative | [56] |
| miR-380-3p | Alpaca melanocytes | SOX6 | Negative | [56] |
| miR-200c | Normal human epidermal keratinocytes (NHEK) | SOX1 | Positive | [69] |
| miR-27a-3p | Alpaca and Mouse melanocytes | Wnt3a | Negative | [54][70] |
| miR-379 | Alpaca melanocytes | IGF1R | Negative | [71] |
| miR-143-5p | Human melanocytes | Myo5a | Negative | [72] |
| miR-143-5p | Alpaca melanocytes | TAK1 | Negative | [73] |
| miR-125b | WM266-4 human melanoma cells, MNT1 human melanoma cells | SH3BP4 | Negative | [60] |
Wu et al. used a miR-434-5p homologue to target TYR in cells cultured in vitro as well as in an animal model and showed efficient melanin synthesis reduction. Similarly, miR-330-5p downregulated TYR in melanoma cells and normal melanocytes, inducing depigmentation without affecting cell proliferation [66]. This miR has been also identified in exosomes derived from keratinocytes [56]. In this study, exosomes carrying miR-330-5p caused a decrease in melanin production and TYR expression in melanocytes. Similarly, miR-330-5p overexpression in melanocytes confirmed its inhibitory activity on melanogenesis. Several other miRNAs, such as miR-203, which targets Kinesin Superfamily Protein 5b, involved in melanosome transfer, and miR-3196, with unknown target genes, have been identified in human-derived exosomes from keratinocytes and have shown an ability to increase melanin content in melanocytes [67]. Indeed, exosomes released by Black keratinocytes, as well as Caucasian UV-irradiated keratinocytes, were able to induce increased TYR activity and melanogenic gene expression in melanocytes, at least partly as a result of miR content.
As reported above, several miRNAs which directly target MITF are involved in melanogenesis. Conversely, MITF expression may be subject to more complex regulation. For example, it has been shown that miR-21a-5p overexpression downregulated the SOX5 target, as well as β-catenin and CDK2 protein expression, in normal human melanocytes. In agreement with SOX5 involvement in melanogenesis, its downregulation induced an increase in MITF and melanogenic enzymes’ expression, with consequent stimulation of melanogenesis [68]. Several other miRNAs have been identified as regulators of SOX transcription factors [56][69][74]. miR-145, which was initially identified as a miR regulated by UV treatment and forskolin in murine melan-a melanocytes, significantly modulates several genes involved in pigmentation. miR-145 overexpression or downregulation reduced or increased the expression of several genes involved in melanin biosynthesis, such as SOX9, MITF, TYR, and TYRP-1 [74][75], and in melanosome transfer, such as Myosin Va (Myo5a), Rab27a, and fascin1 (Fscn1), respectively [76]. Notably, miR-145 targeted the 3′-UTR binding site of Myo5a, a molecular motor involved in the intracellular trafficking of vesicles and organelles. A further miR that binds an SOX family component is miR-380-3p, which targets SOX6 by binding to the 3′-UTR region. In alpaca melanocytes, miR-380-3p overexpression downregulated SOX6 and increased β-catenin. Differently from studies indicating that β-catenin induces MITF transcription, in this study, miR-380-3p overexpression caused decreased mRNA levels of melanin-related genes including MITF, TYR, TYRP-1, and DCT, suggesting other SOX6 activities in MITF regulation [56].
Recently, miR-200c has been identified as a SOX1 regulator. Specifically, miR-200c induced SOX1 downregulation by direct binding to the SOX1 3′-UTR, leading to intranuclear β-catenin upregulation and increased expression of MITF-dependent gene expression as well as melanogenesis. These effects were induced in normal human epidermal melanocytes (NHEM) treated with exosomes containing miR-200c, produced by normal human epidermal keratinocytes (NHEK). Notably, miR-200c was found at low levels in exosomes derived from keratinocytes in vitiligo lesions compared to exosomes from NHEK, with a consequent inhibition in melanin production [69]. Some miRNAs have also been involved in modulating the extracellular ligands which regulate melanogenic signaling pathways. An example is miR-27a-3p, which directly targets Wnt3a, a component of the Wnt signaling pathway. miR-27a-3p was found to be expressed at higher levels in alpaca white skin compared to brown skin [54] and was similarly related to skin color in mice [70]. Furthermore, its functional role in downregulating Wnt3a, and thus inhibiting β-catenin and melanogenesis, was confirmed in mouse melanocytes transfected with miR-27a-3p [70]. In addition, miR-379 was shown to modulate insulin-like growth factor receptor I (IGFR1) in alpaca melanocytes. Insulin-like growth factor 1 (IGF1), mainly produced in dermal cells, has been shown to improve melanogenesis through the cAMP pathway. Likewise, miR-379 overexpression reduced melanogenesis by inhibiting the cAMP response element (CRE)-binding protein (CREB)/(MITF) pathway in alpaca melanocytes [71].
Finally, miRNAs involved in the regulation of melanosome processing and transfer have been also identified. Among them, miR-143-5p targets Myo5a, which belongs to the complex Rab27a/MLPH/Myo5a that connects melanosomes to the actin cytoskeleton in human melanocytes [72]. In a more recent study, Qi S. et al. inhibited miR-143-5p expression using Short Tandem Target Mimics (STTMs) in order to evaluate the functional role of miR-143-5p. As a consequence, increased melanogenic gene expression, including MITF, TYR, and TYRP-1, MLPH, and Rab27, and melanin production were observed [77]. This result indicates the effective possibility of modulating melanin biosynthesis by a miRNA-regulating approach. In addition, miR-143-5p overexpression induces a decrease in TGF-β-activated kinase 1 (TAK1) expression with consequent effects on melanocyte migration and proliferation and MITF downregulation in alpaca melanocytes. Therefore, miR-143-5p may regulate melanogenesis by modulating different gene targets [73]. By means of a bioinformatic approach, followed by experimental validation, miR-125b has also been shown to directly target SRC homology 3 domain-binding protein 4 (SH3BP4), a gene regulated by MITF, which may act by controlling melanogenic enzymes’ distribution to melanosomes or the mTOR signaling pathway. Indeed, when miR-125b was overexpressed in WM266-4 and MNT1 human melanoma cells, decreased levels of the melanogenic enzymes TYR and DCT were observed, although these genes are not direct miR-125b targets [78].
4.3. miRNA Regulated by UV Rays
UV rays can modify intracellular functions in different ways: by directly or indirectly damaging DNA through reactive oxygen species (ROS), inducing apoptosis, cell cycle arrest, and carcinogenesis. The expression profiles of the miRNAs involved in these events are modified by UV irradiation [79][80][81]. Among the UV-induced effects, it is well known that there is a correlation between UVB irradiation and melanogenesis stimulation [1]. miRNA profiling in irradiated mouse melanocytes has been helpful in identifying miRNAs involved in melanogenesis, such as miR-145 [74]. Furthermore, several miRNAs targeting different pigmentation genes have been involved in UV ray effects, including miR-145, miR-137, miR-148, and miR-25 [57][82][83].
miR-340 upregulation was identified in pigmented cells treated with UVB irradiation among miRNAs related to UV irradiation. Studies have shown that miR-340 significantly represses RhoA protein expression and stimulates melanosome transport [81]. Recently, the functional role of miR-340 has been further investigated in immortalized human epidermal melanocytes (Pig-1), confirming that its expression is modulated by UV and that it downregulates MITF expression and melanin synthesis. Notably, miR-340 mimics decreased melanin content in irradiated cells [63]. A further example of a miR potentially being involved in UV-induced pigmentation modulation is miR-21. The latter has been largely investigated in melanoma and shown to be increased by UV radiation in melanocytes, keratinocytes, and fibroblasts [84]. In mouse skin melanocytes, miR-21 has been shown to enhance MITF expression by targeting SOX5 [85]. Inversely, miR-21a-5p has been observed with different levels of activity in UV-irradiated A375.S2 human melanoma and B16F10 melanoma mouse cells. Indeed, in these UV-treated cells, increased melanin content was associated with an increase in α-MSH expression and reduced EGFR and Akt phosphorylation. miR-21 overexpression negatively modulated these UV-induced effects [86]. Interestingly, this miR has been also involved in communication between melanocytes and keratinocytes. Extracellular vesicles derived from UVA-exposed melanocytes modified keratinocyte behavior by inducing miR-21 upregulation and TGF-β and IL-6/STAT3 signaling pathway activation [87]. UVA and UVB rays also induce changes in miRNA produced by keratinocytes [88][89]. Accumulating evidence indicates that exosomes and exosomal miRNAs represent an effective means of communication for melanocytes and keratinocytes [56]. Notably, in Caucasian human keratinocytes, UV rays induce the release of miR, such as hsa-miR-3196, which may stimulate melanin synthesis in melanocytes [67]. Recently, miR-675 production by human keratinocytes has been shown and its role as a paracrine regulator of melanogenesis has been confirmed in vitro, indicating increased exosomal miR-675 in cells irradiated with a 585 nm light emitting diode (LED) and its ability to attenuate melanogenesis in melanocytes [90].
4.4. Common miRNAs in Melanogenesis and Melanomagenesis
Several miRNAs regulating melanogenesis play an important role in melanoma [91]. For instance, miR-340, miR-218, and miR-137, which inhibit melanogenesis, have a tumor suppressor role during melanomagenesis by negatively regulating MITF expression [58][61][64][92]. Other miRNAs which regulate melanogenesis, including miR-379, -200c, -203, -200a-3p, -125b, -183, -508-3p, -675, -143-5p, -141-3p, -145, and -330-5p, can act as tumor suppressors in melanomagenesis, although the target genes may differ from those identified in melanogenesis. On the contrary, miR-21a-5p, -25, and -27a-3p have shown an oncogenic role, being upregulated in metastatic melanoma and during tumor progression [93][94].
Interestingly, some miRNAs involved in both melanogenesis and melanomagenesis, such as miR-21a-3p, -25, -3196, -145, -137, -148, and -675 are induced by UV rays, the main environmental risk factor for melanoma [79][80][81][95].
This entry is adapted from the peer-reviewed paper 10.3390/ijms22116104
References
- D’Orazio, J.; Jarrett, S.; Amaro-Ortiz, A.; Scott, T. UV Radiation and the Skin. Int. J. Mol. Sci. 2013, 14, 2222.
- Miller, A.J.; Mihm, M.C. Melanoma. N. Engl. J. Med. 2006, 355, 51–65.
- Nguyen, N.T.; Fisher, D.E. MITF and UV Responses in Skin: From Pigmentation to Addiction. Pigment. Cell Melanoma Res. 2019, 32, 224–236.
- Hida, T.; Kamiya, T.; Kawakami, A.; Ogino, J.; Sohma, H.; Uhara, H.; Jimbow, K. Elucidation of Melanogenesis Cascade for Identifying Pathophysiology and Therapeutic Approach of Pigmentary Disorders and Melanoma. Int. J. Mol. Sci. 2020, 21, 6129.
- Lee, A.-Y. Recent Progress in Melasma Pathogenesis. Pigment Cell Melanoma Res. 2015, 28, 648–660.
- Fu, C.; Chen, J.; Lu, J.; Yi, L.; Tong, X.; Kang, L.; Pei, S.; Ouyang, Y.; Jiang, L.; Ding, Y.; et al. Roles of Inflammation Factors in Melanogenesis (Review). Mol. Med. Rep. 2020, 21, 1421–1430.
- Pillaiyar, T.; Manickam, M.; Jung, S.-H. Recent Development of Signaling Pathways Inhibitors of Melanogenesis. Cell Signal. 2017, 40, 99–115.
- Madireddy, S.; Crane, J.S. Hypopigmented Macules. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021.
- D’Mello, S.A.N.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Signaling Pathways in Melanogenesis. Int. J. Mol. Sci. 2016, 17, 1144.
- Yamaguchi, Y.; Passeron, T.; Watabe, H.; Yasumoto, K.; Rouzaud, F.; Hoashi, T.; Hearing, V.J. The Effects of Dickkopf 1 on Gene Expression and Wnt Signaling by Melanocytes: Mechanisms Underlying Its Suppression of Melanocyte Function and Proliferation. J. Investig. Dermatol. 2007, 127, 1217–1225.
- Serre, C.; Busuttil, V.; Botto, J.-M. Intrinsic and Extrinsic Regulation of Human Skin Melanogenesis and Pigmentation. Int. J. Cosmet. Sci. 2018, 40, 328–347.
- Horsburgh, S.; Fullard, N.; Roger, M.; Degnan, A.; Todryk, S.; Przyborski, S.; O’Reilly, S. MicroRNAs in the Skin: Role in Development, Homoeostasis and Regeneration. Clin. Sci. 2017, 131, 1923–1940.
- Yamada, T.; Hasegawa, S.; Iwata, Y.; Arima, M.; Kobayashi, T.; Numata, S.; Nakata, S.; Sugiura, K.; Akamatsu, H. UV Irradiation-Induced DNA Hypomethylation around WNT1 Gene: Implications for Solar Lentigines. Exp. Dermatol. 2019, 28, 723–729.
- Jiang, L.; Huang, J.; Hu, Y.; Lei, L.; Ouyang, Y.; Long, Y.; Li, H.; Li, S.; Yang, L.; Yang, Y.; et al. Identification of the CeRNA Networks in α-MSH-Induced Melanogenesis of Melanocytes. Aging 2020, 13, 2700–2726.
- Lanzillotti, C.; De Mattei, M.; Mazziotta, C.; Taraballi, F.; Rotondo, J.C.; Tognon, M.; Martini, F. Long Non-Coding RNAs and MicroRNAs Interplay in Osteogenic Differentiation of Mesenchymal Stem Cells. Front. Cell Dev. Biol. 2021, 9, 646032.
- Slominski, A.; Wortsman, J.; Plonka, P.M.; Schallreuter, K.U.; Paus, R.; Tobin, D.J. Hair Follicle Pigmentation. J. Investig. Dermatol. 2005, 124, 13–21.
- Slominski, A.; Tobin, D.J.; Shibahara, S.; Wortsman, J. Melanin Pigmentation in Mammalian Skin and Its Hormonal Regulation. Physiol. Rev. 2004, 84, 1155–1228.
- Nordlund, J.J. The Melanocyte and the Epidermal Melanin Unit: An Expanded Concept. Dermatol. Clin. 2007, 25, 271–281.
- Schallreuter, K.U.; Kothari, S.; Chavan, B.; Spencer, J.D. Regulation of Melanogenesis—Controversies and New Concepts. Exp. Dermatol. 2008, 17, 395–404.
- Land, E.J.; Ramsden, C.A.; Riley, P.A. Quinone Chemistry and Melanogenesis. Methods Enzym. 2004, 378, 88–109.
- Del Bino, S.; Ito, S.; Sok, J.; Nakanishi, Y.; Bastien, P.; Wakamatsu, K.; Bernerd, F. Chemical Analysis of Constitutive Pigmentation of Human Epidermis Reveals Constant Eumelanin to Pheomelanin Ratio. Pigment Cell Melanoma Res. 2015, 28, 707–717.
- Chang, H.; Choi, H.; Joo, K.-M.; Kim, D.; Lee, T.R. Manassantin B Inhibits Melanosome Transport in Melanocytes by Disrupting the Melanophilin-Myosin Va Interaction. Pigment. Cell Melanoma Res. 2012, 25, 765–772.
- Wakamatsu, K.; Graham, A.; Cook, D.; Thody, A.J. Characterisation of ACTH Peptides in Human Skin and Their Activation of the Melanocortin-1 Receptor. Pigment Cell Res. 1997, 10, 288–297.
- Tagashira, H.; Miyamoto, A.; Kitamura, S.-I.; Tsubata, M.; Yamaguchi, K.; Takagaki, K.; Imokawa, G. UVB Stimulates the Expression of Endothelin B Receptor in Human Melanocytes via a Sequential Activation of the P38/MSK1/CREB/MITF Pathway Which Can Be Interrupted by a French Maritime Pine Bark Extract through a Direct Inactivation of MSK1. PLoS ONE 2015, 10, e0128678.
- Imokawa, G. Melanocyte Activation Mechanisms and Rational Therapeutic Treatments of Solar Lentigos. Int. J. Mol. Sci. 2019, 20, 3666.
- Lin, J.Y.; Fisher, D.E. Melanocyte Biology and Skin Pigmentation. Nature 2007, 445, 843–850.
- Roberts, D.W.; Newton, R.A.; Beaumont, K.A.; Helen Leonard, J.; Sturm, R.A. Quantitative Analysis of MC1R Gene Expression in Human Skin Cell Cultures. Pigment Cell Res. 2006, 19, 76–89.
- Mosca, S.; Cardinali, G.; Flori, E.; Briganti, S.; Bottillo, I.; Mileo, A.M.; Maresca, V. The PI3K Pathway Induced by AMSH Exerts a Negative Feedback on Melanogenesis and Contributes to the Release of Pigment. Pigment Cell Melanoma Res. 2021, 34, 72–88.
- Levy, C.; Khaled, M.; Fisher, D.E. MITF: Master Regulator of Melanocyte Development and Melanoma Oncogene. Trends Mol. Med. 2006, 12, 406–414.
- Vachtenheim, J.; Borovanský, J. “Transcription Physiology” of Pigment Formation in Melanocytes: Central Role of MITF. Exp. Dermatol. 2010, 19, 617–627.
- Ohbayashi, N.; Fukuda, M. Recent Advances in Understanding the Molecular Basis of Melanogenesis in Melanocytes. F1000Research 2020, 9.
- Hsiao, J.J.; Fisher, D.E. The Roles of Microphthalmia-Associated Transcription Factor and Pigmentation in Melanoma. Arch. Biochem. Biophys. 2014, 563, 28–34.
- Khaled, M.; Larribere, L.; Bille, K.; Aberdam, E.; Ortonne, J.-P.; Ballotti, R.; Bertolotto, C. Glycogen Synthase Kinase 3beta Is Activated by CAMP and Plays an Active Role in the Regulation of Melanogenesis. J. Biol. Chem. 2002, 277, 33690–33697.
- Saha, B.; Singh, S.K.; Sarkar, C.; Bera, R.; Ratha, J.; Tobin, D.J.; Bhadra, R. Activation of the Mitf Promoter by Lipid-Stimulated Activation of P38-Stress Signalling to CREB. Pigment Cell Res. 2006, 19, 595–605.
- Wang, Y.; Viennet, C.; Robin, S.; Berthon, J.-Y.; He, L.; Humbert, P. Precise Role of Dermal Fibroblasts on Melanocyte Pigmentation. J. Dermatol. Sci 2017, 88, 159–166.
- Chakraborty, A.K.; Funasaka, Y.; Slominski, A.; Ermak, G.; Hwang, J.; Pawelek, J.M.; Ichihashi, M. Production and Release of Proopiomelanocortin (POMC) Derived Peptides by Human Melanocytes and Keratinocytes in Culture: Regulation by Ultraviolet B. Biochim. Biophys. Acta 1996, 1313, 130–138.
- Cui, R.; Widlund, H.R.; Feige, E.; Lin, J.Y.; Wilensky, D.L.; Igras, V.E.; D’Orazio, J.; Fung, C.Y.; Schanbacher, C.F.; Granter, S.R.; et al. Central Role of P53 in the Suntan Response and Pathologic Hyperpigmentation. Cell 2007, 128, 853–864.
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854.
- Finotti, A.; Fabbri, E.; Lampronti, I.; Gasparello, J.; Borgatti, M.; Gambari, R. MicroRNAs and Long Non-Coding RNAs in Genetic Diseases. Mol. Diagn. Ther. 2019, 23, 155–171.
- Rotondo, J.C.; Mazzoni, E.; Bononi, I.; Tognon, M.; Martini, F. Association Between Simian Virus 40 and Human Tumors. Front. Oncol. 2019, 9, 670.
- Pradhan, A.K.; Bhoopathi, P.; Talukdar, S.; Scheunemann, D.; Sarkar, D.; Cavenee, W.K.; Das, S.K.; Emdad, L.; Fisher, P.B. MDA-7/IL-24 Regulates the MiRNA Processing Enzyme DICER through Downregulation of MITF. Proc. Natl. Acad. Sci. USA 2019, 116, 5687–5692.
- Correia de Sousa, M.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering MiRNAs’ Action through MiRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249.
- Huang, W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. Methods Mol. Biol. 2017, 1617, 57–67.
- Mazziotta, C.; Lanzillotti, C.; Iaquinta, M.R.; Taraballi, F.; Torreggiani, E.; Rotondo, J.C.; Otòn-Gonzalez, L.; Mazzoni, E.; Frontini, F.; Bononi, I.; et al. MicroRNAs Modulate Signaling Pathways in Osteogenic Differentiation of Mesenchymal Stem Cells. Int. J. Mol. Sci. 2021, 22, 2362.
- Iaquinta, M.R.; Lanzillotti, C.; Mazziotta, C.; Bononi, I.; Frontini, F.; Mazzoni, E.; Oton-Gonzalez, L.; Rotondo, J.C.; Torreggiani, E.; Tognon, M.; et al. The Role of MicroRNAs in the Osteogenic and Chondrogenic Differentiation of Mesenchymal Stem Cells and Bone Pathologies. Theranostics 2021, in press.
- Nedaeinia, R.; Manian, M.; Jazayeri, M.H.; Ranjbar, M.; Salehi, R.; Sharifi, M.; Mohaghegh, F.; Goli, M.; Jahednia, S.H.; Avan, A.; et al. Circulating Exosomes and Exosomal MicroRNAs as Biomarkers in Gastrointestinal Cancer. Cancer Gene. Ther. 2017, 24, 48–56.
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402.
- Wawrzyniak, O.; Zarębska, Ż.; Rolle, K.; Gotz-Więckowska, A. Circular and Long Non-Coding RNAs and Their Role in Ophthalmologic Diseases. Acta Biochim. Pol. 2018, 65, 497–508.
- Grillone, K.; Riillo, C.; Scionti, F.; Rocca, R.; Tradigo, G.; Guzzi, P.H.; Alcaro, S.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. Non-Coding RNAs in Cancer: Platforms and Strategies for Investigating the Genomic “Dark Matter”. J. Exp. Clin. Cancer Res. 2020, 39, 117.
- Neagu, M.; Constantin, C.; Cretoiu, S.M.; Zurac, S. MiRNAs in the Diagnosis and Prognosis of Skin Cancer. Front. Cell Dev. Biol. 2020, 8, 71.
- Mione, M.; Bosserhoff, A. MicroRNAs in Melanocyte and Melanoma Biology. Pigment Cell Melanoma Res. 2015, 28, 340–354.
- Itoh, T.; Fukatani, K.; Nakashima, A.; Suzuki, K. MicroRNA-141-3p and MicroRNA-200a-3p Regulate α-Melanocyte Stimulating Hormone-Stimulated Melanogenesis by Directly Targeting Microphthalmia-Associated Transcription Factor. Sci. Rep. 2020, 10, 2149.
- Chen, T.; Zhao, B.; Liu, Y.; Wang, R.; Yang, Y.; Yang, L.; Dong, C. MITF-M Regulates Melanogenesis in Mouse Melanocytes. J. Dermatol. Sci. 2018, 90, 253–262.
- Tian, X.; Jiang, J.; Fan, R.; Wang, H.; Meng, X.; He, X.; He, J.; Li, H.; Geng, J.; Yu, X.; et al. Identification and Characterization of MicroRNAs in White and Brown Alpaca Skin. BMC Genom. 2012, 13, 555.
- Zhu, Z.; He, J.; Jia, X.; Jiang, J.; Bai, R.; Yu, X.; Lv, L.; Fan, R.; He, X.; Geng, J.; et al. MicroRNA-25 Functions in Regulation of Pigmentation by Targeting the Transcription Factor MITF in Alpaca (Lama Pacos) Skin Melanocytes. Domest. Anim. Endocrinol. 2010, 38, 200–209.
- Liu, X.; Du, B.; Zhang, P.; Zhang, J.; Zhu, Z.; Liu, B.; Fan, R. MiR-380-3p Regulates Melanogenesis by Targeting SOX6 in Melanocytes from Alpacas (Vicugna Pacos). BMC Genom. 2019, 20, 962.
- Dong, C.; Wang, H.; Xue, L.; Dong, Y.; Yang, L.; Fan, R.; Yu, X.; Tian, X.; Ma, S.; Smith, G.W. Coat Color Determination by MiR-137 Mediated down-Regulation of Microphthalmia-Associated Transcription Factor in a Mouse Model. RNA 2012, 18, 1679–1686.
- Bemis, L.T.; Chen, R.; Amato, C.M.; Classen, E.H.; Robinson, S.E.; Coffey, D.G.; Erickson, P.F.; Shellman, Y.G.; Robinson, W.A. MicroRNA-137 Targets Microphthalmia-Associated Transcription Factor in Melanoma Cell Lines. Cancer Res. 2008, 68, 1362–1368.
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-Mediated Transfer of MRNAs and MicroRNAs Is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007, 9, 654–659.
- Kim, N.-H.; Choi, S.-H.; Kim, C.-H.; Lee, C.H.; Lee, T.R.; Lee, A.-Y. Reduced MiR-675 in Exosome in H19 RNA-Related Melanogenesis via MITF as a Direct Target. J. Investig. Dermatol. 2014, 134, 1075–1082.
- Guo, J.; Zhang, J.; Wang, W.; Cheung, F.W.; Lu, Y.; Ng, C.; Kung, H.; Liu, W. MicroRNA-218 Inhibits Melanogenesis by Directly Suppressing Microphthalmia-Associated Transcription Factor Expression. RNA Biol. 2014, 11, 732–741.
- Du, B.; Liu, X.; Khan, A.; Wan, S.; Guo, X.; Xue, J.; Fan, R. MiRNA-183∼96∼182 Regulates Melanogenesis, Cell Proliferation and Migration in B16 Cells. Acta Histochem. 2020, 122, 151508.
- Yang, Y.; Wei, X.; Bai, J.; Huang, M.; Hao, T.; Hao, Y.; Wang, Y.; Li, C. MicroRNA-340 Is Involved in Ultraviolet B-Induced Pigmentation by Regulating the MITF/TYRP1 Axis. J. Int. Med. Res. 2020, 48, 300060520971510.
- Goswami, S.; Tarapore, R.S.; Poenitzsch Strong, A.M.; TeSlaa, J.J.; Grinblat, Y.; Setaluri, V.; Spiegelman, V.S. MicroRNA-340-Mediated Degradation of Microphthalmia-Associated Transcription Factor (MITF) MRNA Is Inhibited by Coding Region Determinant-Binding Protein (CRD-BP). J. Biol. Chem. 2015, 290, 384–395.
- Wu, D.T.; Chen, J.S.; Chang, D.C.; Lin, S.-L. Mir-434-5p Mediates Skin Whitening and Lightening. Clin. Cosmet. Investig. Dermatol. 2008, 1, 19–35.
- Rambow, F.; Bechadergue, A.; Saintigny, G.; Morizot, F.; Mahé, C.; Larue, L. MiR-330-5p Targets Tyrosinase and Induces Depigmentation. J. Investig. Dermatol. 2014, 134, 2846–2849.
- Lo Cicero, A.; Delevoye, C.; Gilles-Marsens, F.; Loew, D.; Dingli, F.; Guéré, C.; André, N.; Vié, K.; van Niel, G.; Raposo, G. Exosomes Released by Keratinocytes Modulate Melanocyte Pigmentation. Nat. Commun. 2015, 6, 7506.
- Aguennouz, M.; Guarneri, F.; Oteri, R.; Polito, F.; Giuffrida, R.; Cannavò, S.P. Serum Levels of MiRNA-21-5p in Vitiligo Patients and Effects of MiRNA-21-5p on SOX5, Beta-Catenin, CDK2 and MITF Protein Expression in Normal Human Melanocytes. J. Dermatol. Sci. 2021, 101, 22–29.
- Zhao, C.; Wang, D.; Wang, X.; Mao, Y.; Xu, Z.; Sun, Y.; Mei, X.; Song, J.; Shi, W. Down-Regulation of Exosomal MiR-200c Derived from Keratinocytes in Vitiligo Lesions Suppresses Melanogenesis. J. Cell Mol. Med. 2020, 24, 12164–12175.
- Zhao, Y.; Wang, P.; Meng, J.; Ji, Y.; Xu, D.; Chen, T.; Fan, R.; Yu, X.; Yao, J.; Dong, C. MicroRNA-27a-3p Inhibits Melanogenesis in Mouse Skin Melanocytes by Targeting Wnt3a. Int. J. Mol. Sci. 2015, 16, 921.
- Liu, B.; Zhang, J.; Hu, S.; Qi, S.; Jia, Q.; Yang, W.; Yang, S.; Ji, K.; Liu, X.; Dong, C.; et al. MicroRNA-379 Mediates Pigmentation, Migration and Proliferation of Melanocytes by Targeting the Insulin-like Growth Factor 1 Receptor. Exp. Dermatol. 2020, 29, 467–476.
- Alves, C.P.; Yokoyama, S.; Goedert, L.; Pontes, C.L.S.; Sousa, J.F.; Fisher, D.E.; Espreafico, E.M. MYO5A Gene Is a Target of MITF in Melanocytes. J. Investig. Dermatol. 2017, 137, 985–989.
- Ji, K.; Zhang, P.; Zhang, J.; Fan, R.; Liu, Y.; Yang, S.; Hu, S.; Liu, X.; Dong, C. MicroRNA 143-5p Regulates Alpaca Melanocyte Migration, Proliferation and Melanogenesis. Exp. Dermatol. 2018, 27, 166–171.
- Dynoodt, P.; Mestdagh, P.; Van Peer, G.; Vandesompele, J.; Goossens, K.; Peelman, L.J.; Geusens, B.; Speeckaert, R.M.; Lambert, J.L.W.; Van Gele, M.J.L. Identification of MiR-145 as a Key Regulator of the Pigmentary Process. J. Investig. Dermatol. 2013, 133, 201–209.
- Harris, M.L.; Baxter, L.L.; Loftus, S.K.; Pavan, W.J. Sox Proteins in Melanocyte Development and Melanoma. Pigment Cell Melanoma Res. 2010, 23, 496–513.
- Van Gele, M.; Geusens, B.; Schmitt, A.-M.; Aguilar, L.; Lambert, J. Knockdown of Myosin Va Isoforms by RNAi as a Tool to Block Melanosome Transport in Primary Human Melanocytes. J. Investig. Dermatol. 2008, 128, 2474–2484.
- Qi, S.; Liu, B.; Zhang, J.; Liu, X.; Dong, C.; Fan, R. Knockdown of MicroRNA-143-5p by STTM Technology Affects Eumelanin and Pheomelanin Production in Melanocytes. Mol. Med. Rep. 2019, 20, 2649–2656.
- Kim, K.-H.; Bin, B.-H.; Kim, J.; Dong, S.E.; Park, P.J.; Choi, H.; Kim, B.J.; Yu, S.J.; Kang, H.; Kang, H.H.; et al. Novel Inhibitory Function of MiR-125b in Melanogenesis. Pigment Cell Melanoma Res. 2014, 27, 140–144.
- Jayanthy, A.; Setaluri, V. Light-Regulated MicroRNAs. Photochem. Photobiol. 2015, 91, 163–172.
- Guo, L.; Huang, Z.-X.; Chen, X.-W.; Deng, Q.-K.; Yan, W.; Zhou, M.-J.; Ou, C.-S.; Ding, Z.-H. Differential Expression Profiles of MicroRNAs in NIH3T3 Cells in Response to UVB Irradiation. Photochem. Photobiol. 2009, 85, 765–773.
- Jian, Q.; An, Q.; Zhu, D.; Hui, K.; Liu, Y.; Chi, S.; Li, C. MicroRNA 340 Is Involved in UVB-Induced Dendrite Formation through the Regulation of RhoA Expression in Melanocytes. Mol. Cell Biol. 2014, 34, 3407–3420.
- Jiang, S.; Yu, X.; Dong, C. MiR-137 Affects Melanin Synthesis in Mouse Melanocyte by Repressing the Expression of c-Kit and Tyrp2 in SCF/c-Kit Signaling Pathway. Biosci. Biotechnol. Biochem. 2016, 80, 2115–2121.
- Cha, H.J.; Kim, O.-Y.; Lee, G.T.; Lee, K.S.; Lee, J.H.; Park, I.-C.; Lee, S.-J.; Kim, Y.R.; Ahn, K.J.; An, I.-S.; et al. Identification of Ultraviolet B Radiation-induced MicroRNAs in Normal Human Dermal Papilla Cells. Mol. Med. Rep. 2014, 10, 1663–1670.
- Melnik, B.C.; John, S.M.; Carrera-Bastos, P.; Schmitz, G. MicroRNA-21-Enriched Exosomes as Epigenetic Regulators in Melanomagenesis and Melanoma Progression: The Impact of Western Lifestyle Factors. Cancers 2020, 12, 2111.
- Wang, P.; Zhao, Y.; Fan, R.; Chen, T.; Dong, C. MicroRNA-21a-5p Functions on the Regulation of Melanogenesis by Targeting Sox5 in Mouse Skin Melanocytes. Int. J. Mol. Sci. 2016, 17, 959.
- Lin, K.-Y.; Chen, C.-M.; Lu, C.-Y.; Cheng, C.-Y.; Wu, Y.-H. Regulation of MiR-21 Expression in Human Melanoma via UV-Ray-Induced Melanin Pigmentation. Environ. Toxicol. 2017, 32, 2064–2069.
- Wäster, P.; Eriksson, I.; Vainikka, L.; Öllinger, K. Extracellular Vesicles Released by Melanocytes after UVA Irradiation Promote Intercellular Signaling via MiR21. Pigment Cell Melanoma Res. 2020, 33, 542–555.
- Zhou, B.; Xu, Y.; Permatasari, F.; Liu, W.; Li, W.; Guo, X.; Huang, Q.; Guo, Z.; Luo, D. Characterization of the MiRNA Profile in UVB-Irradiated Normal Human Keratinocytes. Exp. Dermatol. 2012, 21, 317–319.
- Kraemer, A.; Chen, I.-P.; Henning, S.; Faust, A.; Volkmer, B.; Atkinson, M.J.; Moertl, S.; Greinert, R. UVA and UVB Irradiation Differentially Regulate MicroRNA Expression in Human Primary Keratinocytes. PLoS ONE 2013, 8, e83392.
- Jin, S.; Chen, L.; Xu, Z.; Xing, X.; Zhang, C.; Xiang, L. 585 Nm Light-Emitting Diodes Inhibit Melanogenesis through Upregulating H19/MiR-675 Axis in LEDs-Irradiated Keratinocytes by Paracrine Effect. J. Dermatol. Sci. 2020, 98, 102–108.
- Sabarimurugan, S.; Royam, M.M.; Das, A.; Das, S.; Gothandam, K.M.; . Jayaraj, R. Systematic Review and Meta-Analysis of the Prognostic Significance of MiRNAs in Melanoma Patients. Mol. Diagn. Ther. 2018, 22, 653–669.
- Möller, K.; Sigurbjornsdottir, S.; Arnthorsson, A.O.; Pogenberg, V.; Dilshat, R.; Fock, V.; Brynjolfsdottir, S.H.; Bindesboll, C.; Bessadottir, M.; Ogmundsdottir, H.M.; et al. MITF Has a Central Role in Regulating Starvation-Induced Autophagy in Melanoma. Sci. Rep. 2019, 9, 1055.
- Varrone, F.; Caputo, E. The MiRNAs Role in Melanoma and in Its Resistance to Therapy. Int. J. Mol. Sci. 2020, 21, 878.
- Tang, H.; Xu, X.; Xiao, W.; Liao, Y.; Xiao, X.; Li, L.; Li, K.; Jia, X.; Feng, H. Silencing of MicroRNA-27a Facilitates Autophagy and Apoptosis of Melanoma Cells through the Activation of the SYK-dependent MTOR Signaling Pathway. J. Cell. Biochem. 2019, 120, 13262–13274.
- Melnik, B.C. MiR-21: An Environmental Driver of Malignant Melanoma? J. Transl. Med. 2015, 13.
