The derivation of induced pluripotent stem cells (iPSCs) from somatic human cells by Takahashi and Yamanaka in 2007 represented a turning point for the field. For the first time, they provided isogenic pluripotent cells with the potential for personalized cell replacement therapies; no ethical issues would be created by using the somatic cells. This opportunity marks a decisive step compared to the generation of human embryonic stem cells (ESCs) arranged by Thomson et al. in 1998. The production of induced pluripotent stem cells (iPSCs) represent a breakthrough in regenerative medicine, providing new opportunities for understanding basic molecular mechanisms of human development and molecular aspects of degenerative diseases.
1. iPSCs Production
The 2012 Nobel prize sanctioned that specialization of cells is reversible and that adult somatic cells could be reprogrammed to an immature, pluripotent state. However, several years after this breakthrough, the comparison of the characteristics of iPSCs with ESCs made clear that not everything that glitters is gold. Since then, many efforts have been made to better understand the biological peculiarities of iPSCs and make the best use of those cells.
Takahashi and Yamanaka [
1,
3] were able to generate iPSCs using retroviral transduction into adult somatic cells. The process involved four transcription factors: (a) octamer-binding transcription factor 4 (Oct4), (b) sex-determining region Y-box 2 (Sox2), (c) Kruppel Like Factor 4 (KLF4), and (d) the oncogene c-MYC; all four are called OSKM. These were selected after testing many genes supposedly involved in the first stages of ESCs’ development [
3].
Since then, numerous different methods have been established to improve reprogramming efficiency. They included varying the source of cells to be reprogrammed (A), the genes used for the reprogramming, and the methods of reprogramming (B). Takahashi’s retroviral approach reached an efficiency of 0.02% in reprogramming [
3], while further attempts by other groups achieved an efficiency of 0.05–0.08% [
4].
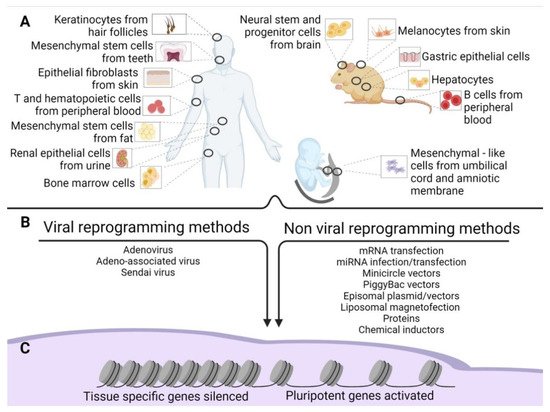
Figure 1. Old and new epigenetic memory in iPSCs. (A) Schematic representation of the possible tissue of origin of the source cells used for reprogramming in human and mouse adult tissues and extraembryonic human tissues. (B) Methods of reprogramming (viral and non-viral) of the source cells. (C) Gene silencing and activation after reprogramming.
These changes in reprogramming methods merged approaches still based on integration into the host genome of exogenous reprogramming factors, and included lentiviruses and transposons as vectors. Those methods offer higher reprogramming efficiency, although the integration process entails permanent DNA modification, potential insertional mutagenesis, and transgenes reactivation.
To overcome these and other potential pitfalls that could limit the future use of iPSCs for tissue regeneration, additional methods were developed based on viral and non-viral, non-integrating delivery of the transgenes containing OSKM [
5,
6,
7,
8,
9,
10]. Further modification of the design of the reprogramming expression vectors and new methods of delivery were designed to minimize or eliminate vector sequences that could be integrated into the iPSCs genome [
11,
12]. However, these different approaches resulted in a significant decrease, in some cases more than 100-fold, of the reprogramming efficiency [
11].
The following sections present the induction methods in two groups: viral and non-viral (B).
1.1. Viral Reprogramming Methods
Within the viral approaches, apart from the aforementioned genome-integrating vectors, we can include Adeno (AV), Adeno-associated (AAV), and Sendai viruses (SV).
Viral non-integrating reprogramming methods were developed to overcome concerns related to exogenous gene integration and DNA modification at the expense of generally lower reprogramming efficiency. For example, the Adenoviral approach permits an efficiency of only 0.001–0.0001% in mouse fibroblasts [
13] and 0.0002% in human fibroblasts [
14]; multiple infections might be required [
15]. In addition, the use of viral vectors might elicit an immune response in the host after cell transplantation, thus compromising the efficacy of the therapy [
16].
1.1.1. Sendai Virus
Sendai virus is a negative sense, mRNA virus belonging to Paramixoviridae family [
17]. It is non-pathogenic to humans, and its use as a viral vector has several advantages: (1) being an mRNA virus, it does not enter the nucleus in its lifecycle, thus eliminating the risk of modifying the host genome and/or causing gene silencing by epigenetic changes [
18]; (2) it shows a broad tropism, being able to infect several cell types in vitro [
19,
20,
21,
22,
23]; (3) due to its non-integrating nature, viral genome is diluted to every cell duplication, allowing its removal from the reprogrammed cells; and (4) it allows the production of a large number of proteins, thus allowing multiplicity of infection (MOI) reduction. Sendai viral vectors were successfully used to reprogram fibroblast cells [
24], as well as blood [
25,
26] and renal epithelial cells in the urine [
27]. This technique is quite efficient, ranging between 0.01% and 4% in the generation of human iPSCs at 25 days of induction [
20,
24,
25,
28,
29]. Up to 10 passages or a high-temperature culture (39 °C) might be necessary to remove the viral genome completely [
11]; however, an auto-erasable, replication-deficient Sendai virus was recently developed using microRNA-302 which impedes viral replication by blocking the viral RNA-dependent RNA polymerase [
30].
1.1.2. Adenovirus
Adenovirus is a non-integrating virus [
31] that remains in the epichromosomal form in all cell types, except in egg cells [
32]. Adenovirus offers a large cargo capacity, a transient expression, and rapid clearance from dividing cells, thus requiring multiple rounds of infection. The reprogramming efficiency is low, 0.001–0.0001% in mouse cells and 0.0002% in human cells, most likely because of the low infection efficiency and the narrow expression window of reprogramming factors [
16].
1.1.3. Adeno-Associated Virus
Adeno-associated virus is a non-pathogenic, non-autonomous single-stranded DNA virus, unable to replicate without the presence of a co-infecting helper virus. In its absence, AAV’s genome remains in episomal form within the infected cells, although integration into the host’s genome was reported in fewer than 10% of cases. For all these reasons, this vector has been used in more than 100 clinical trials [
35]. However, the need for multiple rounds of transduction for cell reprogramming, limited transgene capacity (5 kb), and low efficiency (less than 0.01%) [
16,
35] still limit AAVs’ use as a vector for inducing pluripotency.
1.2. Non-Viral Reprogramming Methods
Along with these techniques, we can include mRNA transfection, miRNA infection/transfection, PiggyBac, Episomal plasmids/vectors, minicircle vectors, and protein and chemical induction [
11,
31].
1.2.1. mRNA Transfection
mRNA transfection was first used for cell reprogramming by Warren’s group. They overcame several obstacles to transcribe mRNAs to express reprogramming factors efficiently, reaching a 1.4% reprogramming efficiency [
36]. Moreover, the addition of Lin28 to the Yamanaka reprogramming factor protocol, valproic acid in the cell culture medium, and a change of O
2 concentration to 5% allowed for an increase in efficiency to 4.4% [
36].
1.2.2. miRNA Infection/Transfection
Regarding the miRNA infection/transfection, several miRNA, such as miR-302b or miR-372, are strongly expressed in ESCs. Their addition to Yamanaka factors increased up to 15-fold reprogramming efficiency for OSKM alone [
37]. Interestingly, some miRNAs could reprogram cells at high efficiency even in the absence of co-transfection with OSKM, bringing the efficiency of reprogramming for BJ-1 fibroblasts up to 10% [
38].
1.2.3. Transposons: PiggyBac and Sleeping Beauty
These two transposons usually consist of a polycistronic transcript containing the OSKM factors joined by 2A peptides, allowing post-translational cleavage of the polyprotein into single reprogramming proteins as well as maintenance of their stoichiometric co-expression [
39]. PiggyBac is a transposon, a mobile genetic element, that can be easily inserted and removed from chromosomal TTAA sites in the genome. Using a transposase, it can be integrated and subsequently excised from the genome [
11]. When the OSKM factors are cloned into the PiggyBac vector and co-transfected into mouse embryonic fibroblasts (MEFs), reprogramming efficiency ranged from 0.02 to 0.05% of the total transfected cells [
40,
41]. This technique requires only a single transfection; the transposon can transport substantial cargo and presents low immunogenicity.
An intrinsic feature of the PiggyBac vector is its integration into the host genome. However, it could be cleanly excised from the iPSCs genome. Potential reintegration is conceivable due to the use of the same enzyme for insertion and excision. This reintegration risk forces a tight screening of iPSC clones to confirm the absence of integration and is time-consuming. As previously mentioned, reprogramming efficiency is quite low (0.01–0.1%) but can be improved using sodium butyrate. Regardless, the efficiency remains 50-fold lower than retroviral-mediated reprogramming methods [
42].
Sleeping Beauty transposon vector differs from PiggyBac in its ability to integrate randomly into host genomes, thus showing no integration tendency with regard to specific genes and gene-regulatory elements [
43].
1.2.4. Episomal Plasmids
Episomal plasmids ensure a technically simple procedure, a stable transgene expression due to their self-replication, and a low immunogenicity, allowing their removal by culturing cells in the absence of drug selection [
44]. Due to their vulnerability to exonucleases, episomal vectors have an extremely low reprogramming efficiency, primarily due to the short expression time in the cells. It is possible to overcome this issue by repeating transfections daily; however, the reprogramming efficiency remains unsatisfactory (0.0003–0.0006%) [
45]. The inclusion of NANOG, LIN28, and LT (SV40 large T gene) as reprogramming factors enhanced the efficiency 100 times, making it comparable to viral-based methods [
46]. Instead of using a single plasmid for every Yamanaka factor, which is laborious and less efficient, as only a few cells receive all the plasmids, some groups use polycystronic plasmids to obtain a “3+1 delivery” of the reprogramming factors (with Oct4, Klf4, and Sox2, carried by one plasmid, and c-Myc by the other) [
15], while other groups rely on one single plasmid to deliver all four genes [
47], under the control of a constitutively active CAG promoter. However, these last methods do not ensure an adequate stoichiometric co-expression of the multiple reprogramming factors. The use of picornaviral 2A self-cleaving peptides to link reprogramming factors, when used as a polycistronic construct [
48], can partially ameliorate the balance of the expression of the four genes. However, there is still the chance that polycistronic plasmid could produce an unbalanced expression of each reprogramming factor [
49]. This issue, together with the large size of the plasmid, hampers the efficiency of plasmid-based reprogramming [
16].
1.2.5. Minicircle Vectors
Minicircle vectors are supercoiled DNA episomal vectors similar to a standard plasmid but containing only the eukaryotic promoter and the cDNA(s) of the genes to be expressed. Their small size, resistance to cleavage, extremely low immunogenicity, and high transfection efficiency make them a good tool for cell reprogramming [
16], despite a very low reprogramming efficiency (0.005%) and long reprogramming time (14–18 days). Thus, multiple rounds of transfection are required, causing a reduction in cell viability [
50]. In order to improve the efficiency of reprogramming, various researchers used electropulsation, included additional reprogramming factors and/or microRNAs, used small molecules, and included hypoxic conditions [
51,
52,
53,
54]. Tight screening of the clones is necessary to exclude the integration of transgene sequences.
1.2.6. Liposomal Magnetofection
Liposomal magnetofection is a non-viral technique that allows the delivery of nucleic acids in cultured cells by mixing nucleic acid and magnetic nanoparticles in cationic lipids. The lipids are concentrated on the surface of the cells using a magnetic field [
55]. This technique has little chance of genomic integration, requires only a single transfection, and has low immunogenicity. There are rare cases of genomic integration. Consequently, screening iPSC clones is necessary to confirm the absence of integration [
56]. Moreover, its reprogramming efficiency is between 0.032% and 0.040% after 8 days [
16].
1.2.7. Proteins
Although the bioactive forms of reprogramming proteins can be synthesized by prokaryotic or eukaryotic systems, the main hurdle for reprogramming is their limited capability to cross the cell membrane. To overcome this obstacle, the protein approach takes advantage of the HIV-TAT protein (a protein transduction domain) in delivering recombinant proteins. This technique allows the introduction of proteins into cells from the external environment without permeabilization agents [
57]. The efficiency of this procedure is quite low, at 0.006% of mouse fibroblasts [
58] and 0.001% of human fibroblasts [
9,
59]. To improve the reprogramming efficiency, some authors supplemented the culture media with valproic acid (VPA), with 0.006% of cells induced to pluripotency after 30–35 days [
58].
Protein transduction domains later became a method to deliver not only proteins but also other macromolecules. Those included peptide nucleic acids (PNA), antisense, short-interfering ribonucleic acids (siRNA), liposomes, iron nanoparticles, and plasmids [
57].
1.2.8. Chemical Induction
Despite the methods mentioned above for reprogramming, the highest degree of safety at the cost of low reprogramming efficiency is represented by iPSC generation through the use of small molecules to obtain chemically induced pluripotent stem cells (CiPSCs).
Hou et al. developed a combination of six small molecules (obtained after an intense work of screening of more than 10,000 compounds). They included several cAMP agonists (Forskolin, Rolipram, and Prostaglandin E2) and epigenetic modulators (sodium butyrate, 3-deazaneplanocin A (DZNep), 5-Azacytidine, and RG108) to generate chemically induced iPSCs (CiPSCs). Interestingly, they found that small molecule (sm) iPSCs could be generated using only one gene of the OSKM, namely Oct4, with the addition of CHIR99021, tranylcypromine (VC6T), VPA, and 616452 [
8,
60]. Compared to ESCs, CiPSCs have similar doubling time, gene expression profiles and differentiation ability, and they generate teratomas and chimeric mice [
8,
61]. Moreover, it is intriguing that different chemical cocktails are needed to induce other source cells [
60]. To date, current chemical reprogramming efficiency is only 0.2%, with an induction time of more than 36 days that was recently reduced to 16 days [
62,
63].
2. Comparison between Different Methods of Reprogramming
The presence of the high level of copy number variation (CNV) in hiPSCs compared to hESCs or human somatic cell samples can be explained with two, not self-excluding, hypotheses: (a) they are gained de novo during the reprogramming procedure or in vitro iPSCs culture or, (b) they are present in the starting somatic cell population that could also be a mosaic [
64,
65,
66]. Since the first work of Yamanaka [
3], many efforts have been made to understand how reprogramming could impact the quality of iPSCs.
In a study conducted by Ma et al., the comparison between different methods of reprogramming (i.e., Sendai virus (IPSCs-S) and retroviral (iPSCs-R) methods) indicated that some lines, such as iPS-S4, iPS-S5, and iPS-R2, did not display significant genomic macroscopic alterations. Copy number variation (CNV) analysis did not entirely exclude the presence of small insertion-deletions (indels), point mutations, or translocations [
67].
In other papers, the genetic stability of independent iPSCs lines with common donors was tested by CNV SNP microarrays [
65]. However, lines produced using integrating vectors showed a trended but not significantly higher frequency of clinically significant CNV (58%) compared with non-integrating vectors (41%). Since this study compared iPSCs lines obtained from the same donor, the authors could evaluate whether the CNV differences were due to the tissue of origin or the method or reprogramming [
65]. Similarly, Schlaeger et al., [
29] compared episomal vector reprogramming, Sendai virus, RNA, and lentivirus reprogramming, finding no differences in CNV. Many different groups found that if differences do exist between the reprogramming methods, these are most likely present when the reprogramming is made using integrating viral vectors; they are also very subtle, although they could be more deleterious [
68,
69,
70,
71].
Taken together, these data suggest that different induction methods do not contribute significantly to the genic alterations found in iPSC lines obtained from isogenic cells; most likely, the genic impairment found in iPSCs could be ascribed to the somatic donor cells or the cultivation time.
3. Source Cells for Reprogramming
The cells used for reprogramming depend on the organism, the availability of the tissue, and the kind of differentiated cells we would like to produce from the iPSCs. As addressed in the following sections, many tissues cannot be used because they are unavailable (i.e., brain tissue) unless obtained as discarded tissue.
However, to date, iPSCs have been obtained using a plethora of tissues (B). In this regard, some of these methods require invasive procedures such as biopsies, as in primary skin fibroblasts. More accessible sources are available, namely peripheral blood from which we can retrieve T cells [
25], B cells [
72], hematopoietic stem cells [
73], and bone marrow cells. Recently, iPSCs have been produced by choosing less invasive cells to obtain, such as keratinocytes isolated from hair follicles [
74]. Very often, cell sources have been obtained from biological waste material [
75]. Examples include renal epithelial cells in the urine [
27,
76], mesenchymal stem cells from teeth and fat tissue [
77], liver and stomach cells [
78], melanocytes [
79], neural stem and progenitor cells [
80], and embryonic and extraembryonic tissue [
81]. These outcomes indicate that cells of all tissues might be converted into iPSCs. The final point about the origin of source cells concerns their age. Senescent cells or cells obtained from the elderly are induced to iPSCs with more difficulties. However, Lapasset et al. found that their induction efficiency could be increased using a six-factor reprogramming cocktail (SOX2, OCT4, KLF4, NANOG, LIN28, and c-MYC) instead of the usual OSKM, which also eliminates the marks of cellular aging [
82,
83].
Depending on the cells of origin and the methods of reprogramming, gene cocktails other than OSKM, such as p53 shRNA, Lin28, L-Myc, SV40LT, Nanog, Glis1, and others, have been used in different reprogramming mixes, sometimes improving the efficiency of reprogramming in particular subsets of tissue sources [
27,
84].
An important aspect that we mentioned earlier is the presence of mosaicisms in the source cells that could negatively affect reprogrammed cells. The production of iPSCs from a patient affected by Down syndrome showed that the patient was a mosaic, since one-third of the reprogrammed cells were euploid, whereas the remaining 66% were trisomic [
85]. Abyzov et al. demonstrated that 50% of the CNVs identified in the hiPSC lines were detectable, even at a very low frequency, in the source fibroblast population [
66], indicating the presence of somatic mosaicism in these cells.
4. Epigenetic Alteration after Reprogramming
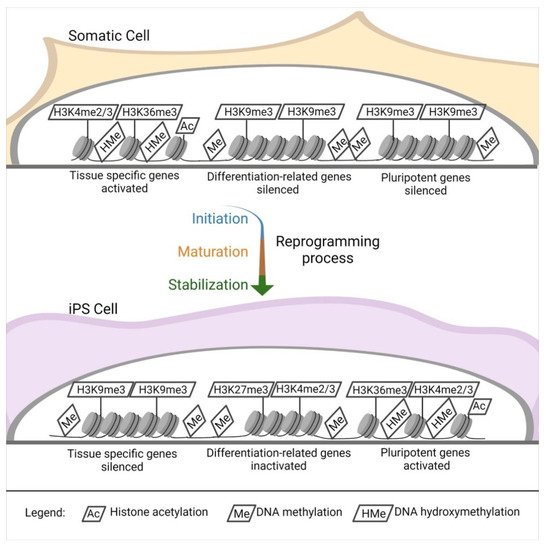
Independently of the reprogramming method, profound modifications of the epigenetic landscape of the donor cells appear during iPSC induction. Pluripotent stem cells [
86] such as ESCs show a distinctive epigenetic profile, with active chromatin modifications. Histone acetylation, hypomethylated DNA, a tri-methylation at the 4th lysine residue of the histone H3 (H3K4me3), and tri-methylation at the 36th lysine residue of the histone H3 (H3K36me3) [
87,
88] locate primarily within the regions of genes responsible for pluripotency ().
Figure 2. Epigenetic landscape changes. DNA and histones modification in the reprogramming process. Starting from a somatic cell throughout the reprogramming process (initiation, maturation, and stabilization), there is an intense modification of the histones and DNA in specific sites.
The opposite happens in tissue-specific genes [
87]. Another interesting aspect of pluripotent stem cells is that they have elevated levels of the so-called bivalent domains, with methylation in H3K27me3 and H3K4me3 in differentiation-related genes, which can be easily activated or silenced, eliminating H3K27me3 or H3K4me3. This sensitive equilibrium is pivotal for the maintenance of stemness [
87,
89,
90] ().
During reprogramming, silencing of somatic cell genes and activation of pluripotency-associated genes are observed (C), and they push for a de-differentiation of the cell into a naïve, pluripotent state. These cells are ultimately characterized by unlimited cell proliferation and differentiation into cells derived from the three germ layers in vitro; in vivo, they can form teratomas, generate chimeras, and complete organisms through tetraploid complementation. These are the most rigorous criteria for pluripotency characterization of pluripotent stem cells [
91], which can be addressed only in non-human cells [
92].
To date, the molecular mechanisms that underlie the derivation and maintenance of iPSCs are not yet wholly understood. Most of the studies on the transcriptional and epigenetic circumstances driving pluripotency and reprogramming have been performed on mice. Due to the strong cross-species similarities, many of these results have been translated to humans.
The change in gene expression occurs progressively due to a defined sequence of cellular and molecular events (). It can be divided into initiation, maturation, and stabilization. Some of the phases concern cellular dynamics in which there is a change in cell size, a mesenchymal-to-epithelial transition, a change in proliferation rate, and a metabolic switch; the stabilization phase is transgene independent. Another aspect concerns transcription dynamics in which the somatic genes are switched off in the initiation phase. At the same time, cell cycle genes are activated from the maturation to the stabilization phase; the pluripotency genes are also activated from the maturation phase. Meanwhile, the epidermis genes are on only in the maturation phase. The epigenetic dynamic H3K4me2 (permissive) and H3K9me3 (repressive) methylation are turned on early in the activation phase and decrease along with the other phases. H3K4me3/H3K27me3 (bivalent) increases from the maturation phase throughout the stabilization phase, similar to DNA hydroxymethylation and histone acetylation. Finally, DNA methylation and demethylation increase during the stabilization phase [
87,
88].
This entry is adapted from the peer-reviewed paper 10.3390/cells10061470