The impact of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) on the world is still expanding. Thus, there is an urgent need to better understand this novel virus and find a way to control its spread. Like other coronaviruses, the nucleocapsid (N) protein is one of the most crucial structural components of SARS-CoV-2. This protein shares 90% homology with the severe acute respiratory syndrome coronavirus N protein, implying functional significance. Based on the evolutionary conservation of the N protein in coronavirus, we reviewed the currently available knowledge regarding the SARS-CoV-2 N protein in terms of structure, biological functions, and clinical application as a drug target or vaccine candidate.
1. Introduction
Coronavirus (CoVs) can cause a variety of diseases in humans and animals, such as infectious gastroenteritis in livestock, infectious bronchitis in chickens, and the common cold with some mild respiratory symptoms in humans [
1,
2]. However, in the past two decades, novel deadly CoVs have emerged, causing three infectious disease pandemics in human society, resulting in enormous health threats and disrupting the global economy [
3]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported in December 2019 and has since spread worldwide, making it the seventh known CoVs to infect humans [
4]. The disease caused by SARS-CoV-2 infection is called COVID-19, and is characterized by symptoms, such as shortness of breath or dyspnea, fever, muscle pain, even death in severe cases [
5]. The cumulative number of confirmed COVID-19 cases already exceeded 200 million worldwide.
Molecular evolution analysis based on nucleic acid sequence alignment shows that SARS-CoV-2 is a member of the genus
β-Coronavirus and the subgenus
Sarbecovirus. The viral genome consists of a positive-sense, single-stranded RNA, which comprises 14 open reading frames (ORFs), encoding 16 nonstructural proteins that make up the replicase complex, nine accessory proteins (ORF), and four structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N) [
4,
6,
7]. Among them, the N protein is highly conserved in the CoVs genus and is one of the most abundant structural proteins in virus-infected cells [
8]. The fundamental function of the N protein is to package the viral genome RNA into a long helical ribonucleocapsid (RNP) complex and to participate in the assembly of the virion through its interactions with the viral genome and membrane protein M [
9]. In addition, the N protein of the CoVs has been shown to be involved in the host cellular machinery such as interferon inhibition, RNA interference, and apoptosis, serving a regulatory role in viral life cycles [
10,
11]. Moreover, the N protein is also an immunodominant antigen in host immune responses that can be used as a diagnostic antigen and immunogen [
12]. As summarized in this article, numerous studies on the N protein of SARS-CoV-2 to investigate the role of the N protein in viral assembly, replication, and the host immune response regulation, to provide a reference for developing specific immune-based drug and vaccine.
2. Composition and Structure of SARS-CoV-2 N Protein
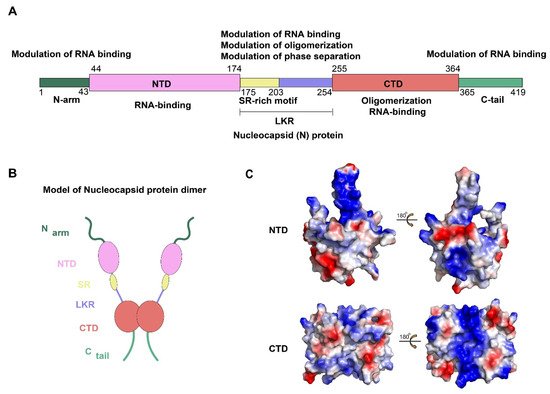
The N protein of SARS-CoV-2 is encoded by the ninth ORF of the virus and is composed of 419 amino acids. Like other CoVs, the SARS-CoV-2 N protein has a modular organization which can be divided into intrinsically disordered regions (IDRs) and conserved structural regions according to the sequence characteristics [
13]. The IDRs include three modules: N-arm, central Ser/Arg-rich flexible linker region (LKR), and C-tail, while the conserved structural regions including two modules: N-terminal domain (NTD) and C-terminal domain (CTD). In the primary structure, NTD and CTD are connected by LKR and are usually flanked by N-arm and C-tail (A,B).
Figure 1. Structural overview of the SARS-CoV-2 N protein. (A,B) Schematic of the SARS-CoV-2 N protein modular organization. The three intrinsically disordered regions, including the N-arm, central Ser/Arg(SR)-rich flexible linker region (LKR) and C-tail, and the N-terminal domain (NTD) and C-terminal domain (CTD) are illustrated. (C) Electrostatic surface of the SARS-CoV-2 N-NTD (PDB ID 6YI3) and N-CTD (PDB ID 7CE0). Blue denotes positive charge potential, while red indicates negative charge potential. All structural figures were prepared using PyMOL.
To date, the NTD and CTD of the SARS-CoV-2 N protein have been solved and show strongly resembles other CoVs N protein structures [
14,
15]. For SARS-CoV-2 N protein, each NTD molecule presents a right-handed fist shape. The core subdomain consists of a five-stranded U-shaped antiparallel β-sheet with the topology β4–β2–β3–β1–β5, sandwiched between two short α-helices (α1 before β2 strand and α2 after β5). There is a large protruding β-hairpin ((β2′–β3′) between β2 and β3 as a bridge to connect them, which stands out of the core (PDB ID: 6YI3). In terms of the CTD, it exists as a tight homodimer and displays an overall rectangular slab shape, in which each protomer is comprised of five α-helices, two β-strands, and two 3
10 helices. The β-hairpin from one protomer is inserted into the cavity of the other protomer, resulting in the formation of the four-stranded, antiparallel β-sheet at the dimer interface. The β sheet forms one face of the slab dimer, while on the opposite face of the dimer, the surface is formed by α-helices and loops (PDB ID: 7CE0). Extensive hydrogen bond interactions between the two hairpins and hydrophobic interactions between the β-sheet and the α-helices make the dimeric structure highly stable [
14]. However, due to various reasons, such as difficulty in maintaining protein stability and the highly disordered sequence of IDRs, there are no structures available for any of the full-length N proteins from CoVs [
16,
17]. Some bioinformatics methods may provide some hints. A recent study compared the IDRs of N protein, as well as other CoVs proteins, between SARS-CoV-2, SARS-CoV, and bat SARS-like CoV, which provide important grounds for a better understanding of the biological functions and structure [
18]. Meanwhile, the information regarding the N-IDRs by using a combination of 2D spectra and nuclear magnetic resonance (NMR) is worthy of consideration [
19].
3. Clinical Applications of the SARS CoV-2 N Protein
3.1. N-Protein as a Diagnostic Marker
Prompt detection is essential to limit the spread of pathogens. The availability of the complete genomic sequence of SARS CoV-2 has facilitated the development of a variety of diagnostic tests for SARS-CoV-2. Reverse transcription polymerase chain reaction (RT-PCR) has been used as a rapid diagnostic test during the epidemic [
52]. However, the sensitivity of viral RNA testing varies depending on the timing of testing relative to exposure, which could lead to false-negative results [
53]. Thus, more and more laboratories pay attention to serological tests.
Among the four CoVs structural proteins, the S and N proteins are the main immunogens [
54]. There have been several serological tests showing that S and N induced a strong antibody response in hosts [
55,
56]. During the detection process, it was found that the detection rate of N protein was higher than that of S protein in PCR-positive patients [
57]. Hence, it is a feasible method to use N proteins for serological tests or combined N and S proteins as capture antigens to increase the sensitivity of this assay. However, one question raised is that the test for specific antibodies against SARS-COV-2 in the serum will appear positive only about 7 days after infection or later, making it difficult to detect the infection at an early stage. Given this situation, it is necessary to explore the diagnostic value of SARS-CoV-2 proteins in the early stages of SARS-COV-2 infection. Several studies have detected serum N protein level in SARS-COV-2 infected patients and analyzed the correlation with serum N protein antibody level using the commercial kit. Based on the CUT-OFF value determined from the receiver operating characteristic (ROC) curve, the specificity of the SARS-COV-2 serum N protein detection was 96.84%, and the sensitivity was 92% before the appearance of antibodies, suggesting that the detection of SARS-COV-2 serum N protein has a high diagnostic value for infected patients before the appearance of antibodies, and shortens the window of serological diagnosis [
58]. Meanwhile, several laboratories have tried to identify the immunodominant epitopes of N protein and develop specific monoclonal antibodies that can be used in ELISA. Amrun et al. identified four immunodominant epitopes: S14P5, S20P2, S21P2, and N4P5, on the S and N viral proteins. IgG responses to all identified epitopes displayed a strong detection profile, with N4P5 achieving the highest level of specificity (100%) and sensitivity (>96%) against SARS-CoV-2, suggesting the feasibility of developing mAbs to these epitopes alone or in combination used in ELISA to detect SARS-CoV-2 [
59].
Taken together, all these data support the notion that the N-protein could be used as an efficient diagnostic tool for detection of SARS-CoV-2 infection and the specific detection methods of N protein should further be validated in more patient samples.
3.2. N Protein: As a Therapeutic Target
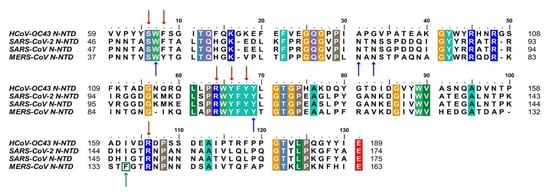
Despite extensive research on COVID-19, there is currently no effective treatment available for clinical use. Based on the conservation of CoVs N protein in evolution and its key role in viral replication, it is a promising target for drug discovery. Firstly, since the RNA binding activity of N protein is pivotal to viral RNP formation and genome replication, blocking the RNA binding of N-NTD has been proven to be a considering strategy. To date, there have been some small compounds targeting other CoVs considered as candidate inhibitors for SARS-CoV-2 by virtual screening. For example, the compounds PJ34 and H3, which targeted the RNA binding site of N-NTD, can inhibit HCoV-OC43 replication [
14]. Notably, the key residues that are involved in the RNA binding interactions, including S51, F53, R107, Y109, Y111, and R149 (in SARS-CoV-2 N-NTD numbering), are conserved, suggesting potential development possibility (1) [
60,
61].
Figure 1. Sequences alignment of four CoVs N-NTD. Multiple sequence alignment of HCoV-OC43 (UniProtKB: P33469), SARS-CoV-2 (UniProtKB: P0DTC9), SARS-CoV (UniProtKB: P59595), MERS-CoV (UniProtKB: K9N4V7). The highly conserved residues were filled with colors. Red arrows indicate conserved RNA binding sites. Blue arrows and green arrow indicate conserved and mutant residues for the non-native interaction interface, respectively. HCoV-OC43, human coronavirus OC43; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus. (The fill color selected in this figure legend is the default setting of the BioEdit software.).
In addition, blocking normal N protein oligomerization or triggering abnormal RNP formation is also an attractive inhibitory strategy. More recently, Lin et al. identified 5-benzyloxygramine (P3) is a novel inhibitor for MERS-CoV by virtual screening. This compound could mediate MERS-CoV N-NTD non-native dimerization and induce N protein aggregation. The structure-based study showed that P3 targets the non-native interface of N-NTD dimers and simultaneously interacts with the hydrophobic pockets in both N-NTD protomers. It was demonstrated that P3 was able to replace the vector-fusion residues of promoter 2 to occupy its binding cavity in promoter 1 under the legend free condition, which, in turn, stabilized the dimeric status by triggering massive hydrophobic interactions [
14,
62]. By comparing the binding sites of P3 in the hydrophobic cavity, it was found that almost all of the residues of the N-NTD involved in the interactions are conserved, except F135 in MERS-CoV, which is replaced by I146 in SARS-CoV-2(). Although both residues are nonpolar amino acids, the effect on SARS-CoV-2 replication needs to be further verified. For other viruses, such as the human immunodeficiency virus and influenza virus, the researchers proposed a strategy to inhibit viral N protein oligomerization by developing competing peptides [
63,
64]. For CoVs, it has been shown that the excessive peptide based on the C-terminal tail sequence can interfere with CTD oligomerization of HCoV-229E N protein and decrease the viral titer, providing a reference for relevant studies on SARS COV-2 N protein [
65]. Notably, the LLPS of N protein induced by viral genomic RNA is also a potential target [
35]. Slowing viral infection by increasing or decreasing the N protein LLPS is a strategy that could be considered. 1,6-hexanediol, lipoic acid, and aminoglycoside kanamycin, each of which potentially alters LLPS by a representative and distinct mechanism. In terms of SARS-CoV-2, further experiments showed that the formation or the size of condensates could be reduced after treatment with these small molecules [
34]. Meanwhile, high-throughput virtual screening is underway, several potential drug candidates have been proposed, and the next focus is on rigorous experimental validation, such as (−)-catechin gallate and (−)-gallocatechin gallate [
66] ().
Table 1. β-CoV inhibitors target N protein.
| Compounds |
Target Domain or Process |
Mechanism |
Reference |
| PJ34, N-(6-oxo-5,6-dihydrophenanthridin-2-yl) (N,N-dimethylamino) acetamide hydrochloride |
NTD |
Reduce RNA binding |
[14,67] |
| H3, 6-chloro-7-(2-morpholin-4-ylethylamino) quinoxaline-5,8-dione |
NTD |
Reduce RNA binding |
[61] |
| (−)-catechin gallate |
NTD |
Reduce RNA binding |
[66] |
| (−)-gallocatechin gallate |
NTD |
Reduce RNA binding |
[66] |
| P3, 5-benzyloxygr- amine |
CTD |
Induce abnormal dimerization |
[62] |
| 1,6-hexanediol |
LLPS |
prevent condensate formation |
[68] |
| Lipoic acid |
LLPS |
Reduce smaller condensate |
[68,6 |
This entry is adapted from the peer-reviewed paper 10.3390/v13061115