Iron–sulfur clusters are essential to almost every life form and utilized for their unique structural and redox-targeted activities within cells during many cellular pathways. Proteins central to the eukaryotic ISC cluster assembly complex include the cysteine desulfurase, a cysteine desulfurase accessory protein, the acyl carrier protein, the scaffold protein and frataxin (in humans, NFS1, ISD11, ACP, ISCU and FXN, respectively). Recent molecular details of this complex (labeled NIAUF from the first letter from each ISC protein outlined earlier), which exists as a dimeric pentamer, have provided real structural insight into how these partner proteins arrange themselves around the cysteine desulfurase, the core dimer of the (NIAUF)2 complex.
- frataxin
- Fe-S cluster biosynthesis
- ISC machinery
1. Introduction
Iron–sulfur (Fe–S) clusters are inorganic cofactors that are essential for cell viability in almost every life form [1]. Essential pathways that utilize Fe–S clusters include, but are not limited to, cellular respiration and ATP production, as well as several aspects related to cell genetics, including DNA synthesis, damage recognition and repair [2]. The redox and structural versatility inherent in Fe–S clusters allows them to actively participate in targeted activities that include electron transfer and chemical activation reactions, provide biomolecular structural integrity, and it allows them to be used as sensors to gauge environmental iron or oxidation levels in cells and cellular compartments [3]. Regardless of the structure, the ubiquitous presence of Fe–S clusters in all of biology highlights their importance in relation to cell viability.
During early anaerobic conditions in the Earth’s atmosphere, the prevalence of iron and sulfur in the environment and ease of their ability to self-assemble led to Fe–S clusters becoming incorporated into evolving biochemical pathways within early cells [4]. Nature responded by evolving tightly controlled enzymatic pathways that protect Fe–S clusters during their assembly; the complexity of these biosynthetic pathways extended to downstream delivery events too, all of which are tightly regulated. Prokaryotes utilize three different bioassembly pathways to produce Fe–S clusters, including the nitrogen fixation pathway (NIF), the sulfur mobilization pathway (SUF) and the iron–sulfur cluster (ISC) assembly pathway [5][6][7]. While having multiple independent assembly pathways in prokaryotes provides these cells with the flexibility to assembly clusters under different environmental conditions, a complete reliance on the mitochondrial ISC pathway in eukaryotes to directly or indirectly assist in producing Fe–S clusters highlights the extreme importance of this pathway.
Housed within the mitochondrial matrix, proteins that are part of the ISC assembly complex utilize ionized iron and sulfur to produce 2Fe–2S clusters [8]. At the core of the ISC multiprotein complex (summarized in [2][9][10]) is the cysteine desulfurase enzyme (NFS1 in humans, Table 1), a pyridoxal phosphate (PLP)-containing enzyme that provides sulfur for Fe–S cluster assembly by cleaving this atom from the side chain of the substrate L-Cysteine and storing it in the form of a persulfide (Figure 1). The scaffold protein (ISCU2 in humans, or as generalized ISCU) receives persulfide sulfur from the NIA complex as well as Fe(II) in a yet identified manner to perform 2Fe–2S cluster assembly. When two copies of each protein combine within the macromolecular structure, these five proteins make up the NIAUF complex that exists as a stable pentameric dimer ((NIAUF)2, referred to simply as NIAUF for the remaining of the review).
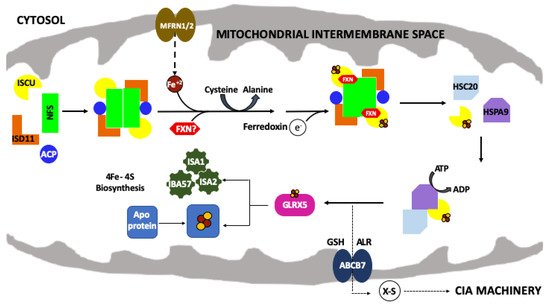
Figure 1. The mitochondrial iron–sulfur cluster (ISC) assembly machinery. Schematic of the de novo mitochondrial Fe–S cluster biosynthetic pathway. Iron (Fe+2) is imported via mitoferrin (MFRN1/2) while cysteine desulfurase (NFS1) provides sulfur, from L-cysteine, in the form of a persulfide (-SSH). ISD11 and ACP stabilize NFS1. The 2Fe–2S cluster is formed on the scaffold protein (ISCU) and ferredoxin (FDX2) provides the electron required for this process. FXN promotes NFS1 activity and Fe loading of ISCU. HSC20, HSPA9 and GLRX5 receive the 2Fe–2S cluster from the ISC complex and promote downstream delivery. The conversion of 2Fe–2S to 4Fe–4S is still uncharacterized but involves a complex of ISA1, ISA2 and IBA57 proteins. The 2Fe–2S cluster is exported out of the mitochondria as an unknown sulfur-containing moiety (X-S) via the ATP binding cassette (ABCB7).
Table 1. Nomenclature of ISC Proteins in the Human, Fly and Yeast Systems.
| Protein Name | Human | Fly | Yeast | |||
|---|---|---|---|---|---|---|
| Symbol | MW (kDa) | Symbol | MW (kDa) | Symbol | MW (kDa) | |
| Cysteine desulfurase | NFS1 | 50.2 | dNfs1 | 51.1 | Nfs1 | 54.5 |
| LYR Motif-containing protein 4 | ISD11 | 10.7 | dIsd11 | 11 | Isd11 | 11.2 |
| Acyl carrier protein | ACP/NDUFAB | 17.4 | dAcp | 17.2 | Acp1 | 13.9 |
| Iron–sulfur cluster assembly scaffold | ISCU | 15.3 | dIscU | 16.7 | Isu1 | 17.8 |
| Frataxin | FXN | 23.1 | dFh | 20.9 | Yfh1 | 19.5 |
The initial structure of the human NI complex core confirmed the surprising presence of E. coli acp, which serendipitously bound to the NFS1/ISD11 complex as a result of the bacterial overexpression system; this initial NIa complex existed in the unique “open” conformation, with the dimeric core of the (NIa)2 structure centered around the two ISD11 proteins at the core dimer interface [11]. This interesting initial structure contrasted with the bacterial cysteine desulfurase dimer observed in a previous bacterial NU report [12]; however, the accessory protein Isd11 is not expressed in prokaryotes. Zinc was known to stabilize the protein’s fold in several early scaffold only structures [13][14][15], and it does so by binding to active site cysteine sulfur ligands. Interestingly, the observation that the NIAUF complex can exist in two dramatically different core orientations (open and closed), and that ISCU exists in two folded states (structured and dynamic), suggests that molecular dynamics likely play an important role in NIAUF function.
Recent reports using the human orthologs, however, indicated that FXN binds to ISCU in an iron-independent manner and that when frataxin, ferredoxin and iron were combined, they were all directly important for Fe(II) loading of ISCU as part of the NIAUFX complex [16]. With regard to scaffold iron loading, the human, fly, yeast and bacterial orthologs were all shown to bind iron at micro to submicromolar affinity at a location on the protein that is distinct from the molecule’s In Isu1, binding iron and zinc does not alter the binding characteristics of the opposing metal, and Zn loaded Isu1 was shown to be functionally inhibited towards Fe–S cluster assembly when part of the NIAUF complex [17]. This is in contrast with recent reports indicating ISCU iron displaces zinc bound to the protein’s active site when FXN is present, and based on this activity it was suggested that Zn loading of ISCU may regulate protein activity and the cluster assembly pathway in vivo [18].
2. The Frataxin Protein
Phenotypes of the disorder include mitochondrial iron overload, breakdown in both heme and Fe–S cluster biosynthesis and subsequent generation of reactive oxygen species that both kill the cell and lead to health complications for the organism [19][20][21][22][23]. Originally thought to participate as a ferritin-like aggregate for mitochondrial iron storage [24], it was later shown that frataxin aggregation activity was dispensable [25] and that frataxin monomers, with a micromolar iron-binding affinity, could directly interact with the scaffold in vitro to promote 2Fe–2S cluster assembly [26]; it was, therefore, suggested that FXN could serve as a chaperone that delivers metal to ISCU [27][28][29]. This idea was supported by in vivo pulldown assays showing the scaffold was the primary binding partner to frataxin in yeast cells and genetic surveys linking frataxin as a direct binding partner [30]. , frataxin is likely iron loaded when in the mitochondrial matrix milieu [26][29][31].
The sequence comparison for the human, fly and yeast orthologs (Figure 2A) shows a high degree of homology (42.8% identity between human and fly, 31.7% identity between the human and yeast) that is maintained with the E. coli bacterial ortholog (24.5% identity between human and bacterial) [32][33][34]. Regions of functional significance, identified by residue color and Roman numerals in Figure 2A and described in detail below, are also highly conserved in function and molecular location. The tertiary structures of the human, fly, and yeast frataxin orthologs are also highly conserved; with members of this family having an α-β sandwich structural motif fold constructed by two large α-helices on one plane of the proteins helical surface and the second surface constructed by six to six β-strands (Figure 2B–D). When viewed together, these proteins have high structural similarities.
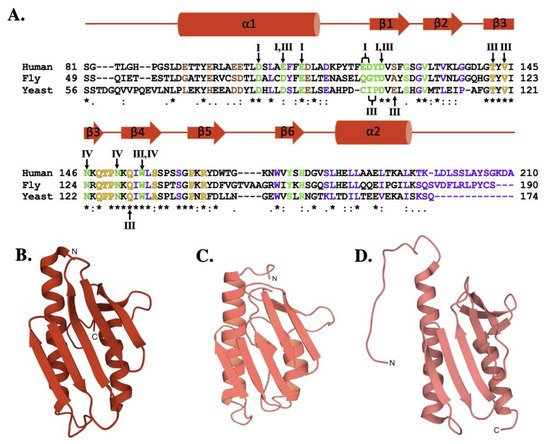
Figure 2. Molecular details of frataxin protein orthologs. (A) Sequence homology between human, fly, and yeast frataxin orthologs. Brown colored letters represent iron-binding residues, green indicates residues interacting with cysteine desulfurase, yellow indicates residues involved in ISCU partnering, and purple indicates residues implicated in stability. Likewise, roman numerals are used to represent residues involved in multiple interactions. Roman numeral I represents iron-binding residues, III indicates residues implicated in protein stability, IV indicates residues involved in ISCU partnering. (B) Crystal structure of human FXN (PDB ID: 1EKG). (C) Crystal structure of fly frataxin modeled using PyMol. (D) Crystal structure of yeast frataxin (PDB ID:2GA5).
The iron-binding properties of frataxin orthologs continue to be of interest related to possible functional activities of the protein during Fe–S cluster bioassembly. As shown by NMR chemical shift perturbation assays, FXN’s iron-binding sites are located at the protein’s N-terminal elements, comprised of acidic side chain atoms from highly conserved Asp and Glu residues both in the protein’s α-helix 1/β-strands 1 and 2 regions (Figure 3A) [16][26][35].
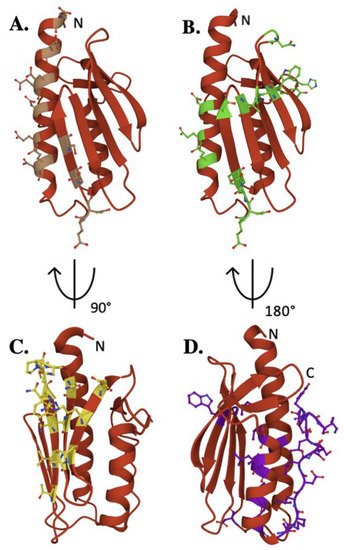
Figure 3. Key residues of biophysical relevance on the human FXN structure. Crystal structure of human FXN with positions of residues for: (A) Fe binding (brown), (B) cysteine desulfurase binding (green), (C) ISCU binding (Yellow) and (D) stability (purple).
Exposed amino acids located at this negatively charged platform provide acidic side chain carboxylate oxygens that can be used for iron binding. Interestingly, human E92 and E96 are not conserved, in contrast to several other iron-binding residues in this region, suggesting their activity could be unique to the human model system. Assuming frataxin iron binding is in part driven by availability of Fe(II) from the mitochondrial matrix free iron pool, the flexibility of these iron-binding sites provide for a dynamic interaction between protein binding partners in an iron-dependent manner. While a direct role for frataxin in binding iron remains unclear, the future development of models to elucidate the NIAUF reaction mechanism should explore a role of frataxin-bound iron in the process.
Frataxin simultaneously binds regions of both the NFS1 proteins within the cysteine desulfurase dimer, interacting primarily with the C-terminal tail of one NFS1 that fastens the complex to ISCU This interaction is further stabilized by hydrogen bonding between residues in the FXN α-helix 1 to the loop and β-strand 1 residues, where human E121, Y123 and D124 form hydrogen bonds with Arg residues in close structural proximity on NFS1 [36][37]. Mutation of W155 impairs ISC biosynthesis and triggers cell death, suggesting this FXN residue plays a key functional role in frataxin’s ability to interact with the active site Cys-loop on NFS1 [36]. Finally, the C-terminus of NFS1 wraps around ISCU and is anchored in place by FXN residues N151, Y175, and H177 [36].
This allows FXN W155 on β-strand 4 to interact with the ISCU active site residues as well as residues on ISCU’s β-strand 3 [36]. An interaction between human FXN W155 and ISCU H137 could cause a conformational change within the complex which stabilizes the NFS1 Cys-loop, providing sulfur transfer to ISCU. A less prominent interface exists from interactions between FXN N151 and the ISCU Ala-loop active site residue C69, capable of causing a conformational change within the Ala-loop [36]. Ultimately, FXN associates with NFS1 to stabilize and position the enzyme for persulfide production and sulfur delivery, while concomitantly forming interactions with ISCU that enable the scaffold to receive the activated sulfur and possibly the Fe(II) necessary for Fe–S cluster production.
The C-terminal region (CTR) of the protein plays a key role in the structural stability of human frataxin. A FXN variant (L198R) was recently identified as a CTR-related FRDA causing mutation [38]. Artificially elongated yeast frataxin is more stable than native protein in yeast, likely because of its ability to fold onto itself in a similar manner as seen in the human protein [39]. The stability and fold of frataxin in the region of the protein’s iron-binding residues is also highly important; disrupting residues in the α-helix 1 region results in decreased frataxin stability and an inability of the protein to bind iron [40][41][42].
3. The Scaffold Protein
ISCU provides the structural architecture on which mitochondrial iron–sulfur cluster assembly is accomplished. A comparison of the human ISCU amino acid sequence with fly and yeast orthologs (Figure 4A, lower) confirms a high-level of conservation between eukaryotic scaffolds (76.0% identity between human and fly, 61.1% between human and yeast) [33][34]. While only the human ISCU structure has been solved, it is likely the high sequence homology translates into a high conservation of secondary structure when compared to the yeast and fly proteins (Figure 4A, upper). The ISCU tertiary structure, determined in complex with the human cysteine desulfurase, consists of four α-helices which arc around the three anti-parallel β-strands to form a planar platform for the molecule (Figure 4B) [36][12]. The ISCU active site is constructed from three conserved cysteines residues and an aspartic acid residue localized at the solvent-exposed protein edge (Figure 4C) [36][15]. The ISCU active site provides the setting where sulfur is delivered as a persulfide by NFS1 with the assistance of FXN, and once iron is delivered both substrates are used for Fe–S cluster assembly [36]. Inability to express a full-length functional ISCU in humans, as observed in the disease ISCU Myopathy [43], results in Fe–S cluster deficiency, emphasizing the scaffold’s importance in relation to the cluster assembly pathway.
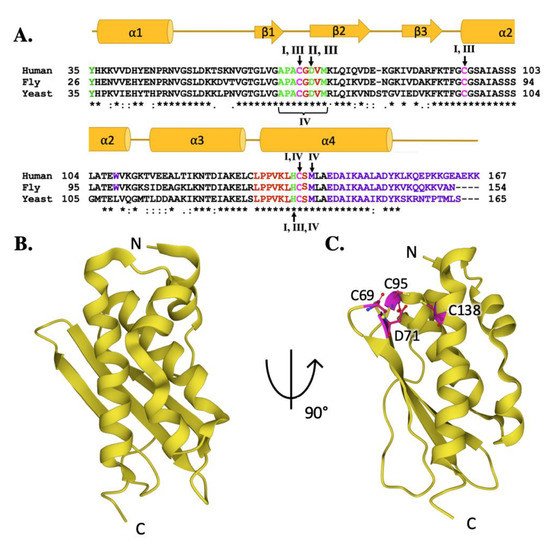
Figure 4. Molecular details of ISCU protein orthologs. (A) Sequence homology between human, fly, and yeast scaffold orthologs. Magenta colored letters indicate active site residues, green indicates residues interacting with cysteine desulfurase, red indicates residues involved in frataxin partnering, and purple indicates residues implicated in stability. Roman numerals are used to represent residues involved in multiple interactions. Roman numeral I represents iron-binding residues, II is active site residues, III indicates residues implicated in protein stability and IV indicates resides involved in frataxin partnering. (B) Crystal structure of human ISCU (PDB ID: 6NZU). (C) Crystal structure of human ISCU (PDB ID: 6NZU) with active site residues highlighted in magenta.
Fe–S cluster assembly is accomplished once the iron and sulfur substrates are delivered, and residues on the scaffold orthologs recognized in the solution and cellular studies to participate in metal binding are highlighted in Figure 5A. In the zinc-loaded human ISCU structures, Zn2+ binds directly to active site residue side chain atoms from C69, D71, C95 and H137 [44][36]. Zinc binding was shown earlier to stabilize the scaffold fold—as a soft acid, it binds to soft base atoms (sulfur) to form a coordination assembly that likely represents binding of a low-spin Fe(II)–S complex [36][45].
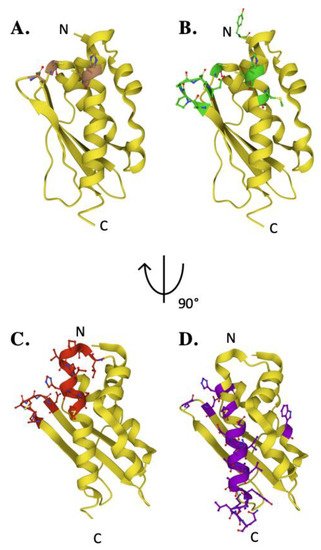
Figure 5. Key residues of biophysical relevance on the human ISCU structure. Crystal structure of human ISCU with positions of residues for: (A) Fe binding (brown), (B) cysteine desulfurase binding (green), (C) FXN binding (red) and (D) stability (purple).
4. Frataxin/Scaffold Interactions
In this section, we analyze the coordinated interactions observed between these two protein partners to help present a comprehensive dynamic picture of how they operate together before, during, and after Fe–S cluster bioassembly. A clear picture of their relative orientation, presented in the recent NIAUF structures, shows the molecular details of how FXN and ISCU interact under controlled conditions (i.e., a structured scaffold protein loaded with Zn2+). It is also beneficial, however, to dissect the additional interactions observed to also occur in solution between these protein partners from the in vivo (genetic analysis) and in vitro (biophysical characterization) analysis, since these interactions add to the overall reaction mechanism story. Beginning with the structural description of the ISCU in complex with NIAU with and without FXN, we will discuss additional observations that frame a dynamic picture of molecular details during assembly by the NIAUF complex.
The interaction observed between FXN and Zn-loaded ISCU, as part of the human NIAUF structure, provides atomic-level insight into the several hydrophilic and hydrophobic intermolecular interactions between conserved residues from both molecules when the ISCU active site is metal loaded with Zn(II) (Figure 6A). Several intermolecular interactions between FXN and ISCU, established through side chain atom contacts between FXN•••ISCU in the structure, include T142•••P133, V144•••V134, W155•••N137 and N151•••C69. FXN also makes contacts with NFS1 through respective FXN•••NFS1 interactions at N146•••A384 and W155•••L386. In this structural orientation, frataxin interacts directly with Zn(II)-loaded ISCU while also interacting with the NFS1 catalytic Cys-loop; in a combined view, this interaction likely would help facilitate sulfur mobilization for liberating the Cys-loop to promote persulfide loading and delivery to ISCU [36][18]. Under both the NIAUF and NIAU structures, ISCU is Zn loaded and the protein’s C-terminal α-helix is completely folded, indicating ISCU in these structures is in its structured conformation.
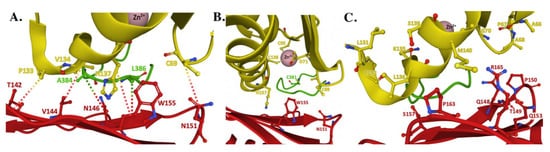
Figure 6. Structure of the human FXN–ISCU interface from the NIAUF crystal structure. (A) Residues with direct interaction on FXN (red) and ISCU (yellow). (B) Residues at the ISCU zinc-loaded active site with the cysteine loop and catalytic residue C381 of NFS1 (green) depicted behind the protein partners. (C) Residues at the FXN–ISCU interface whose association are supported by biochemical data. Adapted from PDB ID:6NZU.
Differences observed in Zn coordination between the NIAU and NIAUF complex structures indicates frataxin alters the zinc-ligand architecture related to NFS1 and ISCU in the cluster assembly active site region of the scaffold. Biochemically, zinc inhibits NFS1 sulfur mobilization in the absence of frataxin, and based on the NIAU structure it does this through direct coordination of Zn(II) to both ISCU active site residues and to the NFS1 catalytic Cys-loop residue C381. so it would be unable to reposition back into the cysteine desulfurase active site pocket for persulfide attachment. Assuming Zn binding to ISCU is physiologically relevant, these structures provide atomic-level details into how zinc could regulate the ISC pathway by controlling the key active site residues in the ISC protein apparatus, as suggested recently [46].
A Zn specific chaperone would be required to load zinc metal onto the scaffold if this were a physiologically relevant process. The affinity for ions, as described by the Irving–Williams series, indicates next to copper, zinc ions have the highest affinity for binding of all the biologically relevant divalent metal ions [47]. Having a weaker binding iron ion displace a bound zinc, especially under basic pH A comparison of the Zn(II) and Fe(II) metal binding affinities for the yeast Isu1 scaffold physically showed zinc binds at an order of magnitude tighter metal binding affinity than iron, so again having Fe displace bound Zn is the opposite of what you would expect.
In the NIAUF structure, interface contacts between frataxin and scaffold are dominated by exposed residues on the FXN β-sheet surface and on the ISCU α-helix 4. From the perspective of frataxin, residues identified and marked with respect to the human sequence including Q148, T149, P150, Q153 and S157 in the β-strand 3-loop-β-strand 4 region along with P163 and R165 on β-strand 5, all of which are on FXN’s β-sheet surface, have also been implicated as interacting with scaffold orthologs (Figure 7C). In the case of the scaffold orthologs, relevant residues on the human ISCU sequence including A66-A68 and G70 in the β-strand 1-loop-β-strand 2 region as well as α-helix 4 residues L131, P132, K135, L139 and M140 are reported to make interactions with frataxin in solution. To allow for these interactions to occur, there would need to be a shift in the relative binding orientation between FXN and ISCU from what is reported in the NIAUF structure, or an unfolding on the ISCU N-terminal region at α-helix 4, which would make this region flexible enough to interact with FXN in a manner not represented in the current structure.
5. Health Relevance
There are so many exciting labs providing new details related to how the ISC protein complex assembles and functions, and their molecular and atomic details provided by several structural biology groups, helps enrich our knowledge of the pathway. However, this work also raises additional important questions. These questions include, how is iron introduced and used by the ISC machinery for Fe–S cluster assembly? Does ISCU bind as part of the NIAUF complex pre-loaded with Fe(II), and if so, does it require a yet unknown iron chaperone to load its metal or is iron be abstracted from the abundant mitochondrial free iron pool [48]?
There are also questions at the atomic level that need to be addressed when developing a comprehensive Fe–S cluster reaction mechanism model for protein activity of the ISC pathway. What is the role of the ISCU active site residues in coordinating metal loading and in assembling 2Fe–2S clusters? 2Fe–2S loaded structure serves as a direct cluster binding ligand in this structure, so is this residue utilized to direct Fe(II) ligation when forming the cluster? to early- or late-stage events that occur during Fe–S cluster assembly?
Getting answers to these either high-level or narrowly focused questions will help the Fe–S cluster assembly community build a more comprehensive understanding of events that lead to ISC protein complex reactivity. When considerations associated with the FXN–ISCU intermolecular interactions that are not obvious from the current NIAUF complex structures are clarified, or considerations related to the scaffold dynamics (as a monomer or part of the complex) are addressed, we will have a more dynamic model of specific events that allow for the production of these essential Fe-cofactors in eukaryotes.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22116006
References
- Braymer, J.J.; Freibert, S.A.; Rakwalska-Bange, M.; Lill, R. Mechanistic concepts of iron-sulfur protein biogenesis in Biology. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118863.
- Lill, R.; Freibert, S.A. Mechanisms of Mitochondrial Iron-Sulfur Protein Biogenesis. Annu. Rev. Biochem. 2020, 89, 471–499.
- Mettert, E.L.; Kiley, P.J. How Is Fe-S Cluster Formation Regulated? Annu. Rev. Microbiol. 2015, 69, 505–526.
- Fassbinder, J.W.; Stanjek, H.; Vali, H. Occurrence of magnetic bacteria in soil. Nature 1990, 343, 161–163.
- Baussier, C.; Fakroun, S.; Aubert, C.; Dubrac, S.; Mandin, P.; Py, B.; Barras, F. Making iron-sulfur cluster: Structure, regulation and evolution of the bacterial ISC system. Adv. Microb. Physiol. 2020, 76, 1–39.
- Garcia, P.S.; Gribaldo, S.; Py, B.; Barras, F. The SUF system: An ABC ATPase-dependent protein complex with a role in Fe-S cluster biogenesis. Res. Microbiol. 2019, 170, 426–434.
- Outten, F.W. Recent advances in the Suf Fe-S cluster biogenesis pathway: Beyond the Proteobacteria. Biochim. Biophys. Acta 2015, 1853, 1464–1469.
- Schilke, B.; Voisine, C.; Beinert, H.; Craig, E. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomydes cervisiae. Proc. Natl. Acad. Sci. USA 1999, 96, 10206–10211.
- Ciofi-Baffoni, S.; Nasta, V.; Banci, L. Protein networks in the maturation of human iron-sulfur proteins. Metallomics 2018, 10, 49–72.
- Maio, N.; Jain, A.; Rouault, T.A. Mammalian iron-sulfur cluster biogenesis: Recent insights into the roles of frataxin, acyl carrier protein and ATPase-mediated transfer to recipient proteins. Curr. Opin. Chem. Biol. 2020, 55, 34–44.
- Cory, S.A.; Van Vranken, J.G.; Brignole, E.J.; Patra, S.; Winge, D.R.; Drennan, C.L.; Rutter, J.; Barondeau, D.P. Structure of human Fe-S assembly subcomplex reveals unexpected cysteine desulfurase architecture and acyl-ACP-ISD11 interactions. Proc. Natl. Acad. Sci. USA 2017, 114, E5325–E5334.
- Shi, R.; Proteau, A.; Villarroya, M.; Moukadiri, I.; Zhang, L.; Trempe, J.F.; Matte, A.; Armengod, M.E.; Cygler, M. Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 2010, 8, 1–18.
- Iannuzzi, C.; Adrover, M.; Puglisi, R.; Yan, R.; Temussi, P.A.; Pastore, A. The role of zinc in the stability of the marginally stable IscU scaffold protein. Protein Sci. Publ. Protein Soc. 2014, 23, 1208–1219.
- Liu, J.; Oganesyan, N.; Shin, D.H.; Jancarik, J.; Yokota, H.; Kim, R.; Kim, S.H. Structural characterization of an iron-sulfur cluster assembly protein IscU in a zinc-bound form. Proteins 2005, 59, 875–881.
- Ramelot, T.A.; Cort, J.R.; Goldsmith-Fischman, S.; Kornhaber, G.J.; Xiao, R.; Shastry, R.; Acton, T.B.; Honig, B.; Montelione, G.T.; Kennedy, M.A. Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site. J. Mol. Biol. 2004, 344, 567–583.
- Cai, K.; Frederick, R.O.; Tonelli, M.; Markley, J.L. Interactions of iron-bound frataxin with ISCU and ferredoxin on the cysteine desulfurase complex leading to Fe-S cluster assembly. J. Inorg. Biochem. 2018, 183, 107–116.
- Lewis, B.E.; Mason, Z.; Rodrigues, A.V.; Nuth, M.; Dizin, E.; Cowan, J.A.; Stemmler, T.L. Unique roles of iron and zinc binding to the yeast Fe-S cluster scaffold assembly protein “Isu1”. Metallomics 2019, 11, 1820–1835.
- Gervason, S.; Larkem, D.; Mansour, A.B.; Botzanowski, T.; Muller, C.S.; Pecqueur, L.; Le Pavec, G.; Delaunay-Moisan, A.; Brun, O.; Agramunt, J.; et al. Physiologically relevant reconstitution of iron-sulfur cluster biosynthesis uncovers persulfide-processing functions of ferredoxin-2 and frataxin. Nat. Commun. 2019, 10, 3566.
- Muhlenhoff, U.; Richhardt, N.; Ristow, M.; Kispal, G.; Lill, R. The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum. Mol. Genet. 2002, 11, 2025–2036.
- Rotig, A.; de Lonlay, P.; Chretien, D.; Foury, F.; Koenig, M.; Sidi, D.; Munnich, A.; Rustin, P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat. Genet. 1997, 17, 215–217.
- Puccio, H.; Simon, D.; Cossee, M.; Criqui-Filipe, P.; Tiziano, F.; Melki, J.; Hindelang, C.; Matyas, R.; Rustin, P.; Koenig, M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet. 2001, 27, 181–186.
- Chen, O.S.; Hemenway, S.; Kaplan, J. Inhibition of Fe-S cluster biosynthesis decreases mitochondrial iron export: Evidence that Yfh1p affects Fe-S cluster synthesis. Proc. Natl. Acad. Sci. USA 2002, 99, 12321–12326.
- Duby, G.; Foury, F.; Ramazzotti, A.; Herrmann, J.; Lutz, T. A non-essential function for yeast frataxin in iron-sulfur cluster assembly. Hum. Mol. Genet. 2002, 11, 2635–2643.
- Cavadini, P.; O’Neill, H.A.; Benada, O.; Isaya, G. Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Hum. Mol. Genet. 2002, 11, 217–227.
- Aloria, K.; Schilke, B.; Andrew, A.; Craig, E.A. Iron-induced oligomerization of yeast frataxin homologue Yfh1 is dispensable in vivo. EMBO Rep. 2004, 5, 1096–1101.
- Cook, J.D.; Bencze, K.Z.; Jankovic, A.D.; Crater, A.K.; Busch, C.N.; Bradley, P.B.; Stemmler, A.J.; Spaller, M.R.; Stemmler, T.L. Monomeric Yeast Frataxin Is an Iron-Binding Protein. Biochemistry 2006, 45, 7767–7777.
- Kondapalli, K.C.; Kok, N.M.; Dancis, A.; Stemmler, T.L. Drosophila Frataxin: An Iron Chaperone during Cellular Fe−S Cluster Bioassembly. Biochemistry 2008, 47, 6917–6927.
- Subramanian, P.; Rodrigues, A.V.; Ghimire-Rijal, S.; Stemmler, T.L. Iron chaperones for mitochondrial Fe-S cluster biosynthesis and ferritin iron storage. Curr. Opin. Chem. Biol. 2011, 15, 312–318.
- Yoon, T.; Cowan, J.A. Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 2003, 125, 6078–6084.
- Wang, T.; Craig, E.A. Binding of yeast frataxin to the scaffold for Fe-S cluster biogenesis, Isu. J. Biol. Chem. 2008, 283, 12674–12679.
- Bou-Abdallah, F.; Adinolfi, S.; Pastore, A.; Laue, T.M.; Dennis Chasteen, N. Iron binding and oxidation kinetics in frataxin CyaY of Escherichia coli. J. Mol. Biol. 2004, 341, 605–615.
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.; Britto, R.; Bursteinas, B.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020, 49.
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 1–6.
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145.
- Huang, J.; Dizin, E.; Cowan, J.A. Mapping iron binding sites on human frataxin: Implications for cluster assembly on the ISU Fe-S cluster scaffold protein. J. Biol. Inorg. Chem. 2008, 13, 825–836.
- Fox, N.G.; Yu, X.; Feng, X.; Bailey, H.J.; Martelli, A.; Nabhan, J.F.; Strain-Damerell, C.; Bulawa, C.; Yue, W.W.; Han, S. Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism. Nat. Commun. 2019, 10, 2210.
- Schmucker, S.; Martelli, A.; Colin, F.; Page, A.; Wattenhoffer-Donze, M.; Puccio, H. Mammalian Frataxin: An Essential Function for Cellular Viability through an Interaction with a Preformed ISCU/NFS1/ISD11 Iron-Sulfur Assembly Complex. PLoS ONE 2011, 6, e16199.
- Faraj, S.E.; Roman, E.A.; Aran, M.; Gallo, M.; Santos, J. The alteration of the C-terminal region of human frataxin distorts is structural dynamics and function. FEBS J. 2014, 281, 3397–3419.
- Adinolfi, S.; Nair, M.; Politou, A.; Bayer, E.; Martin, S.; Temussi, P.; Pastore, A. The Factors Governing the Thermal Stability of Frataxin Orthologues: How To Increase a Protein’s Stability. Biochemistry 2004, 43, 6511–6518.
- Campuzano, V.; Montermini, L.; Molto, M.D.; Pianese, L.; Cossee, M.; Cavalcanti, F.; Monros, E.; Rodius, F.; Duclos, F.; Monticelli, A.; et al. Friedreich’s ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 1996, 271, 1423–1427.
- Clark, E.; Butler, J.S.; Isaacs, C.J.; Napierala, M.; Lynch, D.R. Selected missense mutations impair frataxin processing in Friedreich ataxia. Ann. Clin. Transl. Neurol. 2017, 4, 575–584.
- Petrosino, M.; Pasqup, A.; Novak, L.; Toto, A.; Gianni, S.; Mantuano, E.; Veneziano, L.; Minicozzi, V.; Pastore, A.; Puglisi, R.; et al. Characterization of human frataxin missense variants in cancer tissues. Hum. Mutat. 2019, 40, 1400–1413.
- Mochel, F.; Knight, M.A.; Tong, W.H.; Hernandez, D.; Ayyad, K.; Taivassalo, T.; Andersen, P.M.; Singleton, A.; Rouault, T.A.; Fischbeck, K.H.; et al. Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am. J. Hum. Genet. 2008, 82, 652–660.
- Boniecki, M.T.; Freibert, S.A.; Muhlenhoff, U.; Lill, R.; Cygler, M. Structure and functional dynamics of the mitochondrial Fe/S cluster synthesis complex. Nat. Commun. 2017, 8, 1287.
- Becker, E.M.; Greer, J.M.; Ponka, P.; Richardson, D.R. Erythroid differentiation and protoporphyrin IX down-regulate frataxin expression in Friend cells: Characterization of frataxin expression compared to molecules involved in iron metabolism and hemoglobinization. Blood 2002, 99, 3813–3822.
- Srour, B.; Gervason, S.; Monfort, B.; D’Autreaux, B. Mechanism of Iron-Slufur Cluster Assembly, In the Intimacy of Iron and SUlfur Encounter. Inorganics 2020, 8, 55.
- Irving, H.M.N.H.; Williams, R.J.P. The stability of transition-metal complexes. J. Chem. Soc. 1953, 75, 3192–3210.
- Holmes-Hampton, G.P.; Miao, R.; Garber Morales, J.; Guo, Y.; Munck, E.; Lindahl, P.A. A nonheme high-spin ferrous pool in mitochondria isolated from fermenting Saccharomyces cerevisiae. Biochemistry 2010, 49, 4227–4234.
