Spontaneous tumors can develop in different organs of various plant species without any pathogen infection and, as a rule, appear in plants with a certain genotype: Mutants, interspecific hybrids, etc. In particular, among the inbred lines of radish (Raphanus sativus L.), lines that form spontaneous tumors on the taproot during the flowering period were obtained many years ago. In this work, we analyzed the differential gene expression in the spontaneous tumors of radish versus the lateral roots using the RNA-seq method. Data were obtained indicating the increased expression of genes associated with cell division and growth (especially genes that regulate G2-M transition and cytokinesis) in the spontaneous tumor. Among genes downregulated in the tumor tissue, genes participating in the response to stress and wounding, mainly involved in the biosynthesis of jasmonic acid and glucosinolates, were enriched. Our data will help elucidate the mechanisms of spontaneous tumor development in higher plants.
- Raphanus sativus
- spontaneous tumor
- RNA-seq
- differential gene expression
- cell division
- stress response
1. Introduction
The tumor (syn.: Neoplasm) is a pathological structure emerging as a result of uncontrolled proliferation of a group of cells leaving the systemic control of growth rate, cell differentiation, and proliferation. Therefore, the elucidation of the mechanisms of tumor formation may help identify the key regulators of systemic mechanisms controlling cell proliferation and differentiation. Tumor-like structures are found in almost all multicellular organisms, including higher plants. Pathogen-induced tumors, which make up the majority of neoplasms in higher plants, develop under the influence of infectious agents (bacteria, viruses, fungi, nematodes, insects, etc.), which create a niche for their own habitation in the host plant’s organism mostly by shifting the phytohormonal balance and sometimes activating the meristematic competence of plant cells or modulation of the plant cell cycle. At the same time, much rarer spontaneous tumors of higher plants are formed in plants with certain genotypes (mutants, interspecific hybrids, inbred lines) in the absence of any pathogen, which make them closer to animal tumors [1].
The object of our research is spontaneous tumorigenesis in inbred lines of radish (Raphanus sativus var. radicula Pers.). The genetic collection of radish was created in St. Petersburg State University by selfing individual plants of four cultivars [2], and now it contains 32 highly inbred lines [3]. Eleven radish lines of different origins are characterized by the spontaneous formation of tumors on the roots of plants at the beginning of the flowering stage [3] (Figure 1). Spontaneous tumors in radish originate from the pericycle and cambium cells [4][5], which brings them closer to the lateral and adventitious roots. Moreover, young growing tumors contain meristematic foci in the periphery, which resemble root apical meristems due to the presence of indole-3-acetic acid (IAA) response maxima and RsWOX5 expression [5]. The genetic analysis revealed that the spontaneous tumor formation in radish is a polygenic trait, however, it is also inherited as a monogenic recessive trait in several hybrid combinations [6]. As with most examples of spontaneous tumors in plants, the exact cause of spontaneous tumor formation in radish inbred lines is unknown, but the most likely reason is associated with an increase of free cytokinins (CKs) content, which was found in the roots of tumor-producing radish lines [6].
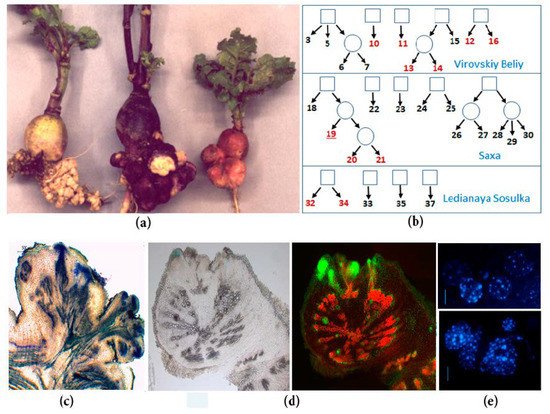
Figure 1. Spontaneous tumors of radish inbred lines: (a) Spontaneous tumors on the taproots of different inbred lines of SPbU genetic collection of Raphanus sativus (from left to right: Lines 34, 16, and 19); (b) the origin of inbred radish lines of SPbU genetic collection. Lines with spontaneous tumor formation are marked in red; (c) anatomy of mature spontaneous tumors in radish: Mass of undifferentiated cells in the periphery of tumor, vascularization of the proximal part of tumor, and the connection of tumor and plant vascular systems; (d) analysis of cell proliferation intensity in the radish taproot with young tumors: Cells with active DNA synthesis were incorporated EdU (5-ethynyl-2’-deoxyuridine) and fluorescently labeled with Alexa Fluor-488 (right) [5]; (e) cytological analysis of roots and tumors in radish inbred line 19 (DAPI staining): Tumor cells (below) have an increased number of chromocenters, which indicate an increased level of ploidy in comparison with taproot cells (above) [7].
In the present study, we used RNA-seq to analyze the changes in gene expression at the early stages of development of spontaneous tumors in radish inbred line 19, while the lateral roots of the same plants were used as the control. The data we obtained revealed the differential expression of more than 1600 genes. We performed the analysis of GO, KEGG, and GSEA categories and also individual transcripts and revealed the upregulation of genes of pathways associated with cell division and expansion, while among the downregulated, the genes involved in stress response were overrepresented. Our findings were confirmed with qPCR for selected transcripts. Since the examples of spontaneous tumors in plants are very rare, and the mechanisms of their development are poorly understood, our study contributes to the understanding of the genetic control of spontaneous tumors development in radish inbred lines in particular and in higher plants in general.
2. Development and Findings
Tumors of higher plants are insufficiently studied, but represent a very interesting phenomenon, since they arise due to the release of a population of cells out of a very reliably arranged systemic control of the proliferation of plant cells. Spontaneous tumors in higher plants are especially rare, and the exact mechanisms of development of most of them remain unclear. In recent years, technologies of next-generation sequencing (NGS) have proven to be instrumental for studying various biological phenomena in different plants [8][9]. RNA sequencing (RNA-seq) is used to assess the functional state of biological tissue based on the variety and abundance of RNA molecules in the sample, so RNA-seq makes it possible to identify candidates for the role of regulators of plant tumor formation. To date, RNA-seq approaches were used for studying the gene expression during the development of only one type of plant tumor, the crown gall caused by A. tumefaciens [10][11]. In our work, we used RNA-seq to analyze the changes in gene expression in the spontaneous tumor of the radish inbred line compared with the lateral root of the same line of radish.
We obtained data demonstrating the significant difference between spontaneous tumors and lateral roots in gene expression profiles: both by pathways and by individual transcripts. The data that seemed most interesting to us were also verified by qPCR. In particular, in the spontaneous tumors, we observed a significant activation of genes associated with the control of cell division: among the pathways, the most enriched were those associated with the control of the cell cycle, DNA replication, cytokinesis, and cell growth. Among numerous upregulated genes involved in cell division, the genes controlling the processes of the G2-M transition and cytokinesis were most widely represented. Moreover, among them there were radish homologs of genes encoding the central participants in these processes in Arabidopsis, such as genes of CDKBs and its interacting partners, B-class cyclins, which provide the G2-M transition [12], as well as such important regulators of cytokinesis as centromere-associated histone H3 variant CEN3, which represents the assembly site for the kinetochore complex of active centromeres [13] or syntaxin KNOLLE, which is essential for phragmoplast formation [14]. The upregulation of genes involved in the control of the cell cycle and cytokinesis reflects the high frequency of cell divisions in spontaneous tumors (Figure 1). In addition, among the upregulated “cell division genes”, there were also inhibitors of CDKs, an increase in the expression of which is usually associated with an increase in the frequency of endomitoses, e.g., during the formation of one of the types of plant tumors—root galls induced by nematodes [15][16]. Indeed, in the spontaneous radish tumors, a high level of polyploid cells was noted compared to normal root tissues (Figure 1). Finally, in our early studies using qPCR, we revealed an increase in the expression levels of genes associated with cell division, including cell cycle genes and some of meristem regulators, in the spontaneous tumors of radish [5][17]. Thus, data on the upregulation of genes involved in the control of cell division in spontaneous tumors of radish are not unexpected for us.
At the same time, the pathways associated with the response to biotic and abiotic stresses were mostly downregulated in the spontaneous tumors. Among genes involved in these pathways, the most widely represented were genes controlling the biosynthesis of JA and the response to them, as well as genes that regulate the synthesis of specific secondary metabolites that are synthesized when plants are wounded or eaten by insects, for example, glucosinolates and lignin [18][19]. Thus, among downregulated genes, there were radish homologs of the Arabidopsis genes that regulate all stages of the JA biosynthesis from the beginning until the end [20], as well as genes that regulate the biosynthesis of glucosinolates [21] and common precursors of glucosinolates and IAA [22][23], and biosynthesis of lignin [24].
These data were also not very unexpected, since the intensification of cell division can lead to tissue juvenilization. The work of age-dependent resistance mechanisms leads to a gain or reinforcement of disease resistance in mature plant organs, whereas juvenile organs with a high level of cell divisions are more susceptible to pathogens [25]. In the adult plant, the activation of age-dependent resistance takes place under the control of genes involved in the regulation of flowering, such as LEAFY [25]. Therefore, the downregulation of genes involved in the flowering control which was revealed in the tumor tissue, may also contribute to the suppression of the age-dependent resistance in the tumor.
The results on changes in the expression of genes that regulate the level of CK and GA and the response to them were quite unexpected for us: based on the data obtained, the balance of these hormones in tumors should sharply shift towards GA. CK and GA are known to antagonistically interact in the control of meristem development, where CK promotes cell division, and GA is responsible for cell growth and differentiation in the periphery [26][27]. Therefore, one should rather expect the activation of CK biosynthesis and suppression of GA biosynthesis in the tumor. Moreover, our previous data indicate the sharp increase of free CK content in the spontaneous tumor of radish [6], and CK were shown to induce tumor formation in radish in vitro [4][28]. It is possible that the downregulation of “CK genes” and an upregulation of “GA genes” revealed by RNA-seq was the result of the work of hypothetical negative feedbacks that reduce the hyperactivation of CK biosynthesis in the radish tumor.
Finally, in the spontaneous tumors, the downregulation of genes that are involved in the control of photomorphogenesis and flowering was revealed. These data were rather unexpected and we can explain the expression of flowering-associated genes in the radish root only by suggesting the broader functions of these genes, e.g., in the radish storage root development. On the other hand, it was reported that individual circadian clock regulators can also participate in providing the plant stress response and affecting SA and JA signaling [29][30]. Therefore, these photomorphogenesis- and flowering-associated genes may be downregulated in the tumor together with other stress-related genes.
Thus, for the first time, data were obtained, which reflect changes in the expression of various gene families in spontaneous tumors in radish lines. A further study of the functions of revealed up- and downregulated genes will help elucidate the mechanisms of plant tumors development.
This entry is adapted from the peer-reviewed paper 10.3390/plants10050919
References
- Dodueva, I.E.; Lebedeva, M.A.; Kuznetsova, K.A.; Gancheva, M.S.; Paponova, S.S.; Lutova, L.L. Plant Tumors: A Hundred Years of Study. Planta 2020, 251, 82.
- Narbut, S.I. Genetic tumor in a radish obtained by inbreeding. Vestn. Leningr. Univ. Biol. 1967, 3, 144–149.
- Buzovkina, I.S.; Lutova, L.A. The Genetic Collection of Radish Inbred Lines: History and Prospects. Russ. J. Genet. 2007, 43, 1181–1192.
- Ilina, E.; Dodueva, I.; Ivanova, N.; Lutova, L. The Effect of Cytokinins on in Vitro Cultured Inbred Lines of Raphanus sativus Var. Radicula Pers. with Genetically Determined Tumorigenesis. Russ. J. Plant Physiol. 2006, 53, 514–522.
- Lebedeva Osipova, M.A.; Tvorogova, V.E.; Vinogradova, A.P.; Gancheva, M.S.; Azarakhsh, M.; Ilina, E.L.; Demchenko, K.N.; Dodueva, I.E.; Lutova, L.A. Initiation of Spontaneous Tumors in Radish (Raphanus sativus): Cellular, Molecular and Physiological Events. J. Plant Physiol. 2015, 173, 97–104.
- Matveeva, T.V.; Frolova, N.V.; Smets, R.; Dodueva, I.E.; Buzovkina, I.S.; Van Onckelen, H.; Lutova, L.A. Hormonal Control of Tumor Formation in Radish. J. Plant Growth Regul. 2004, 23, 37–43.
- Betekhtin, A.; Siwińska, D.; Hasterok, R.; Dodueva, I.; Ilina, E.; Lutova, L. Cytological and Histological Analysis of Radish Root Tumors. In Proceedings of the VII Meeting of Russian Society of Plant Physiologists, International Conference “Plant Physiology—Fundamentals of Ecology and Biotechnology Innovation”, Nizhny Novgorod, Russia, 4–10 July 2011.
- Hearn, D.J.; O’Brien, P.; Poulsen, T.M. Comparative Transcriptomics Reveals Shared Gene Expression Changes during Independent Evolutionary Origins of Stem and Hypocotyl/Root Tubers in Brassica (Brassicaceae). PLoS ONE 2018, 13, e0197166.
- McNeil, M.D.; Bhuiyan, S.A.; Berkman, P.J.; Croft, B.J.; Aitken, K.S. Analysis of the Resistance Mechanisms in Sugarcane during Sporisorium Scitamineum Infection Using RNA-Seq and Microscopy. PLoS ONE 2018, 13, e0197840.
- Tkachenko, A.A.; Gancheva, M.S.; Tvorogova, V.E.; Danilov, L.G.; Predeus, A.V.; Dodueva, I.E.; Lutova, L.L. Transcriptome Analysis of Crown Gall in Radish (Raphanus sativus L.) Inbred Lines. Ann. Appl. Biol. 2021, 178, 527–548.
- Willig, C.J.; Duan, K.; Zhang, Z.J. Transcriptome Profiling of Plant Genes in Response to Agrobacterium Tumefaciens-Mediated Transformation. Curr. Top. Microbiol. Immunol. 2018, 418, 319–348.
- Weimer, A.K.; Biedermann, S.; Harashima, H.; Roodbarkelari, F.; Takahashi, N.; Foreman, J.; Guan, Y.; Pochon, G.; Heese, M.; Van Damme, D.; et al. The Plant-Specific CDKB1-CYCB1 Complex Mediates Homologous Recombination Repair in Arabidopsis. EMBO J. 2016, 35, 2068–2086.
- Lermontova, I.; Rutten, T.; Schubert, I. Deposition, Turnover, and Release of CENH3 at Arabidopsis Centromeres. Chromosoma 2011, 120, 633–640.
- Touihri, S.; Knöll, C.; Stierhof, Y.-D.; Müller, I.; Mayer, U.; Jürgens, G. Functional Anatomy of the Arabidopsis Cytokinesis-Specific Syntaxin KNOLLE. Plant J. Cell Mol. Biol. 2011, 68, 755–764.
- De Almeida Engler, J.; Gheysen, G. Nematode-Induced Endoreduplication in Plant Host Cells: Why and How? Mol. Plant Microbe Interact. 2012, 26, 17–24.
- Cabral, D.; Banora, M.Y.; Antonino, J.D.; Rodiuc, N.; Vieira, P.; Coelho, R.R.; Chevalier, C.; Eekhout, T.; Engler, G.; Veylder, L.D.; et al. The Plant WEE1 Kinase Is Involved in Checkpoint Control Activation in Nematode-Induced Galls. New Phytol. 2020, 225, 430–447.
- Lutova, L.A.; Dodueva, I.E. Role of Meristem-Specific Genes of Plants in Formation of Genetic Tumors. Russ. J. Dev. Biol. 2007, 38, 350–362.
- Johansson, O.N.; Fantozzi, E.; Fahlberg, P.; Nilsson, A.K.; Buhot, N.; Tör, M.; Andersson, M.X. Role of the Penetration-Resistance Genes PEN1, PEN2 and PEN3 in the Hypersensitive Response and Race-Specific Resistance in Arabidopsis Thaliana. Plant J. 2014, 79, 466–476.
- Kumar, D. Salicylic Acid Signaling in Disease Resistance. Plant Sci. 2014, 228, 127–134.
- Wasternack, C.; Strnad, M. Jasmonates Are Signals in the Biosynthesis of Secondary Metabolites—Pathways, Transcription Factors and Applied Aspects—A Brief Review. New Biotechnol. 2019, 48, 1–11.
- Blažević, I.; Montaut, S.; Burčul, F.; Olsen, C.E.; Burow, M.; Rollin, P.; Agerbirk, N. Glucosinolate Structural Diversity, Identification, Chemical Synthesis and Metabolism in Plants. Phytochemistry 2020, 169, 112100.
- Reintanz, B.; Lehnen, M.; Reichelt, M.; Gershenzon, J.; Kowalczyk, M.; Sandberg, G.; Godde, M.; Uhl, R.; Palme, K. Bus, a Bushy Arabidopsis CYP79F1 Knockout Mutant with Abolished Synthesis of Short-Chain Aliphatic Glucosinolates. Plant Cell 2001, 13, 351–367.
- Mikkelsen, M.D.; Naur, P.; Halkier, B.A. Arabidopsis Mutants in the C-S Lyase of Glucosinolate Biosynthesis Establish a Critical Role for Indole-3-Acetaldoxime in Auxin Homeostasis. Plant J. Cell Mol. Biol. 2004, 37, 770–777.
- Sattler, S.E.; Funnell-Harris, D.L. Modifying Lignin to Improve Bioenergy Feedstocks: Strengthening the Barrier against Pathogens? Front. Plant Sci. 2013, 4, 70.
- Hu, L.; Yang, L. Time to Fight: Molecular Mechanisms of Age-Related Resistance. Phytopathology 2019, 109, 1500–1508.
- Greenboim-Wainberg, Y.; Maymon, I.; Borochov, R.; Alvarez, J.; Olszewski, N.; Ori, N.; Eshed, Y.; Weiss, D. Cross Talk between Gibberellin and Cytokinin: The Arabidopsis GA Response Inhibitor SPINDLY Plays a Positive Role in Cytokinin Signaling. Plant Cell 2005, 17, 92–102.
- Fleishon, S.; Shani, E.; Ori, N.; Weiss, D. Negative Reciprocal Interactions between Gibberellin and Cytokinin in Tomato. New Phytol. 2011, 190, 609–617.
- Lutova, L.; Dodueva, I. Genetic Control of Regeneration Processes of Radish Plants In Vitro: From Phenotype to Genotype. Biol. Commun. 2019, 64, 124–132.
- Zhang, C.; Gao, M.; Seitz, N.C.; Angel, W.; Hallworth, A.; Wiratan, L.; Darwish, O.; Alkharouf, N.; Dawit, T.; Lin, D.; et al. LUX ARRHYTHMO Mediates Crosstalk between the Circadian Clock and Defense in Arabidopsis. Nat. Commun. 2019, 10, 2543.
- Yamaura, S.; Yamauchi, Y.; Makihara, M.; Yamashino, T.; Ishikawa, A. CCA1 and LHY Contribute to Nonhost Resistance to Pyricularia Oryzae (Syn. Magnaporthe Oryzae) in Arabidopsis Thaliana. Biosci. Biotechnol. Biochem. 2020, 84, 76–84.
