Coronavirus disease (COVID-19) caused by the SARS-CoV-2 virus has been affecting the world since the end of 2019. The severity of the disease can range from an asymptomatic or mild course to acute respiratory distress syndrome (ARDS) with respiratory failure, which may lead to death. Since the outbreak of the pandemic, scientists around the world have been studying the genome and molecular mechanisms of SARS-CoV-2 infection to develop effective therapies and prevention. In this review, we summarize the progressive development of various treatments and vaccines as they have emerged, a year after the outbreak of the pandemic. Initially for COVID-19, patients were recommended drugs with presumed antiviral, anti-inflammatory, and antimicrobial effects that were previously used to treat other diseases. Thereafter, therapeutic interventions were supplemented with promising approaches based on antibodies, peptides, and stem cells.
- SARS-CoV-2
- COVID-19
- cytokine storm
- therapy
- antibodies
- vaccines
1. COVID-19 Outbreak
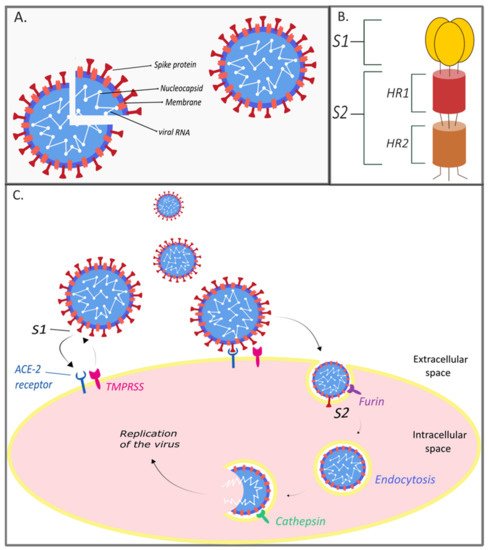
2. The Genome and Structure of SARS-CoV-2
3. Vaccine Development
This entry is adapted from the peer-reviewed paper 10.3390/pathogens10060636
References
- Lai, C.-C.; Shih, T.-P.; Ko, W.-C.; Tang, H.-J.; Hsueh, P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924.
- Cascella, M.; Rajnik, M.; Cuomo, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation, and Treatment of Coronavirus. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020.
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506.
- Wacharapluesadee, S.; Tan, C.W.; Maneeorn, P.; Duengkae, P.; Zhu, F.; Joyjinda, Y.; Kaewpom, T.; Chia, W.N.; Ampoot, W.; Lim, B.L.; et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 2021, 12, 972.
- Chagla, Z.; Hota, S.; Khan, S.; Mertz, D. International Hospital and Community Epidemiology Group Airborne Transmission of COVID-19. Clin. Infect. Dis. 2020.
- Klompas, M.; Baker, M.A.; Rhee, C. Airborne Transmission of SARS-CoV-2: Theoretical Considerations and Available Evidence. JAMA 2020, 4, 15.
- Chen, W.; Lan, Y.; Yuan, X.; Deng, X.; Li, Y.; Cai, X.; Li, L.; He, R.; Tan, Y.; Deng, X.; et al. DetecTable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg. Microbes Infect. 2020, 9, 469–473.
- Cheung, K.S.; Hung, I.F.N.; Chan, P.P.Y.; Lung, K.C.; Tso, E.; Liu, R.; Ng, Y.Y.; Chu, M.Y.; Chung, T.W.H.; Tam, A.R.; et al. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples From a Hong Kong Cohort: Systematic Review and Meta-analysis. Gastroenterology 2020, 159, 81–95.
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844.
- Della Gatta, A.N.; Rizzo, R.; Pilu, G.; Simonazzi, G. Coronavirus disease 2019 during pregnancy: A systematic review of reported cases. Am. J. Obs. Gynecol. 2020, 223, 36–41.
- Yan, J.; Guo, J.; Fan, C.; Juan, J.; Yu, X.; Li, J.; Feng, L.; Li, C.; Chen, H.; Qiao, Y.; et al. Coronavirus disease 2019 in pregnant women: A report based on 116 cases. Am. J. Obs. Gynecol. 2020, 223, 111.e1–111.e14.
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513.
- Esakandari, H.; Nabi-Afjadi, M.; Fakkari-Afjadi, J.; Farahmandian, N.; Miresmaeili, S.-M.; Bahreini, E. A comprehensive review of COVID-19 characteristics. Biol. Proced. Online 2020, 22, 19.
- Docherty, A.B.; Harrison, E.M.; Green, C.A.; Hardwick, H.E.; Pius, R.; Norman, L.; Holden, K.A.; Read, J.M.; Dondelinger, F.; Carson, G.; et al. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: Prospective observational cohort study. BMJ 2020, 369, 1985.
- Aziz, M.; Perisetti, A.; Lee-Smith, W.M.; Gajendran, M.; Bansal, P.; Goyal, H. Taste Changes (Dysgeusia) in COVID-19: A Systematic Review and Meta-analysis. Gastroenterology 2020, 159, 1132–1133.
- Meng, X.; Deng, Y.; Dai, Z.; Meng, Z. COVID-19 and anosmia: A review based on up-to-date knowledge. Am. J. Otolaryngol. 2020, 41, 102581.
- Zheng, J.; Wong, L.-Y.R.; Li, K.; Verma, A.K.; Ortiz, M.E.; Wohlford-Lenane, C.; Leidinger, M.R.; Knudson, C.M.; Meyerholz, D.K.; McCray, P.B.; et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 2021, 589, 603–607.
- Weiss, S.R.; Leibowitz, J.L. Coronavirus Pathogenesis. Adv. Virus Res. 2011, 81, 85–164.
- Chan, J.F.-W.; To, K.K.-W.; Tse, H.; Jin, D.-Y.; Yuen, K.-Y. Interspecies transmission and emergence of novel viruses: Lessons from bats and birds. Trends Microbiol. 2013, 21, 544–555.
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes. Infect. 2020, 9, 221–236.
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. Severe acute respiratory syndrome-related coronavirus: The species and its viruses—A statement of the Coronavirus Study Group. bioRxiv 2020, 7, 937862.
- Wang, H.; Li, X.; Li, T.; Zhang, S.; Wang, L.; Wu, X.; Liu, J. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 24, 1–7.
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574.
- Tang, Q.; Song, Y.; Shi, M.; Cheng, Y.; Zhang, W.; Xia, X.-Q. Inferring the hosts of coronavirus using dual statistical models based on nucleotide composition. Sci. Rep. 2015, 5, 17155.
- Song, Z.; Xu, Y.; Bao, L.; Zhang, L.; Yu, P.; Qu, Y.; Zhu, H.; Zhao, W.; Han, Y.; Qin, C. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses 2019, 11, 59.
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell. Host Microbe 2020, 27, 325–328.
- Wu, C.; Zheng, M.; Yang, Y.; Gu, X.; Yang, K.; Li, M.; Liu, Y.; Zhang, Q.; Zhang, P.; Wang, Y.; et al. Furin: A Potential Therapeutic Target for COVID-19. iScience 2020, 23, 101642.
- Hayashi, T.; Ura, T.; Abiko, K.; Mandan, M.; Yaegashi, N.; Konishi, I. Reasons why new coronavirus, SARS-CoV-2 infections are likely to spread. J. Genet. Med. Gene Ther. 2020, 3, 1–3.
- Zhu, Z.; Zhang, Z.; Chen, W.; Cai, Z.; Ge, X.; Zhu, H.; Jiang, T.; Tan, W.; Peng, Y. Predicting the receptor-binding domain usage of the coronavirus based on kmer frequency on spike protein. Infect. Genet. Evol. 2018, 61, 183–184.
- Du, L.; Zhao, G.; He, Y.; Guo, Y.; Zheng, B.-J.; Jiang, S.; Zhou, Y. Receptor-binding domain of SARS-CoV spike protein induces long-term protective immunity in an animal model. Vaccine 2007, 25, 2832–2838.
- Datta, P.K.; Liu, F.; Fischer, T.; Rappaport, J.; Qin, X. SARS-CoV-2 pandemic and research gaps: Understanding SARS-CoV-2 interaction with the ACE2 receptor and implications for therapy. Theranostics 2020, 10, 7448–7464.
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5.
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8.
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734.
- Wang, Q.; Qiu, Y.; Li, J.-Y.; Zhou, Z.-J.; Liao, C.-H.; Ge, X.-Y. A Unique Protease Cleavage Site Predicted in the Spike Protein of the Novel Pneumonia Coronavirus (2019-nCoV) Potentially Related to Viral Transmissibility. Virol. Sin. 2020, 19, 1–3.
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020, 41, 1100–1115.
- Fontanet, A.; Autran, B.; Lina, B.; Kieny, M.P.; Karim, S.S.A.; Sridhar, D. SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet 2021, 397, 952–954.
- McNally, A. What makes new variants of SARS-CoV-2 concerning is not where they come from, but the mutations they contain. BMJ 2021, 372, 504.
- Burki, T. Understanding variants of SARS-CoV-2. Lancet 2021, 397, 462.
- Prompetchara, E.; Ketloy, C.; Palaga, T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020, 38, 1–9.
- Dimitrova, E.K. Comirnaty. Available online: (accessed on 20 March 2021).
- Glanville, D. EMA Recommends COVID-19 Vaccine Moderna for Authorisation in the EU. Available online: (accessed on 20 March 2021).
- Wang, F.; Kream, R.M.; Stefano, G.B. An Evidence Based Perspective on mRNA-SARS-CoV-2 Vaccine Development. Med. Sci. Monit. 2020, 26, e924700.1-8.
- WHO. W.H.O. Update37-Vaccine-Development.pdf. Available online: (accessed on 9 January 2021).
- Reichmuth, A.M.; Oberli, M.A.; Jaklenec, A.; Langer, R.; Blankschtein, D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv 2016, 7, 319–334.
- Dimitrova, E.K. EMA Starts Second Rolling Review of a COVID-19 Vaccine. Available online: (accessed on 12 January 2021).
- Pinho, A.C. EMA Recommends COVID-19 Vaccine AstraZeneca for Authorisation in the EU. Available online: (accessed on 30 March 2021).
- Van Doremalen, N.; Lambe, T.; Spencer, A.; Belij-Rammerstorfer, S.; Purushotham, J.N.; Port, J.R.; Avanzato, V.; Bushmaker, T.; Flaxman, A.; Ulaszewska, M.; et al. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 2020, 586, 578–582.
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020, 396, 467–478.
- Ramasamy, M.N.; Minassian, A.M.; Ewer, K.J.; Flaxman, A.L.; Folegatti, P.M.; Owens, D.R.; Voysey, M.; Aley, P.K.; Angus, B.; Babbage, G.; et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): A single-blind, randomised, controlled, phase 2/3 trial. Lancet 2020, 396, 1979–1993.
- Watanabe, Y.; Mendonça, L.; Allen, E.R.; Howe, A.; Lee, M.; Allen, J.D.; Chawla, H.; Pulido, D.; Donnellan, F.; Davies, H.; et al. Native-like SARS-CoV-2 spike glycoprotein expressed by ChAdOx1 nCoV-19/AZD1222 vaccine. bioRxiv 2021, 42, 6463.
- Greinacher, A.; Thiele, T.; Warkentin, T.E.; Weisser, K.; Kyrle, P.A.; Eichinger, S. Thrombotic Thrombocytopenia after ChAdOx1 nCov-19 Vaccination. N. Engl. J. Med. 2021.
- Scully, M.; Singh, D.; Lown, R.; Poles, A.; Solomon, T.; Levi, M.; Goldblatt, D.; Kotoucek, P.; Thomas, W.; Lester, W. Pathologic Antibodies to Platelet Factor 4 after ChAdOx1 nCoV-19 Vaccination. N. Engl. J. Med. 2021.
- Francisco, E.M. COVID-19 Vaccine Janssen. Available online: (accessed on 20 March 2021).
- Pinho, A.C. EMA Starts Rolling Review of the Sputnik V COVID-19 Vaccine. Available online: (accessed on 20 March 2021).
- RAPS COVID-19 Vaccine Tracker. Available online: (accessed on 20 March 2021).
- Logunov, D.Y.; Dolzhikova, I.V.; Shcheblyakov, D.V.; Tukhvatulin, A.I.; Zubkova, O.V.; Dzharullaeva, A.S.; Kovyrshina, A.V.; Lubenets, N.L.; Grousova, D.M.; Erokhova, A.S.; et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: An interim analysis of a randomised controlled phase 3 trial in Russia. Lancet 2021, 397, 671–681.
- Thiagarajan, K. What do we know about India’s Covaxin vaccine? BMJ 2021, 373, 997.
- Mallapaty, S. China’s COVID vaccines are going global—But questions remain. Nature 2021, 4, 47.
- Mahase, E. Covid-19: Novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ 2021, 372, 296.
- Waissengrin, B.; Agbarya, A.; Safadi, E.; Padova, H.; Wolf, I. Short-term safety of the BNT162b2 mRNA COVID-19 vaccine in patients with cancer treated with immune checkpoint inhibitors. Lancet Oncol. 2021, 22, 581–583.
- Gavriatopoulou, M.; Ntanasis-Stathopoulos, I.; Korompoki, E.; Terpos, E.; Dimopoulos, M.A. SARS-CoV-2 Vaccines in Patients with Multiple Myeloma. Hemasphere 2021, 5, e547.
- Salmerón Ríos, S.; Mas Romero, M.; Cortés Zamora, E.B.; Tabernero Sahuquillo, M.T.; Romero Rizos, L.; Sánchez-Jurado, P.M.; Sánchez-Nievas, G.; Señalada, J.J.B.; García Nogueras, I.; Estrella Cazalla, J.d.D.; et al. Immunogenicity of the BNT162b2 vaccine in frail or disabled nursing home residents: COVID-A study. J. Am. Geriatr. Soc. 2021, 2, 41.
- Bird, S.; Panopoulou, A.; Shea, R.L.; Tsui, M.; Saso, R.; Sud, A.; West, S.; Smith, K.; Barwood, J.; Kaczmarek, E.; et al. Response to first vaccination against SARS-CoV-2 in patients with multiple myeloma. Lancet Haematol. 2021, 7, 19.
- Terpos, E.; Trougakos, I.P.; Gavriatopoulou, M.; Papassotiriou, I.; Sklirou, A.D.; Ntanasis-Stathopoulos, I.; Papanagnou, E.-D.D.; Fotiou, D.; Kastritis, E.; Dimopoulos, M.A. Low Neutralizing Antibody Responses Against SARS-CoV-2 in Elderly Myeloma Patients After the First BNT162b2 Vaccine Dose. Blood 2021, 5, 16.
