Adaptive immunity relies on the V(D)J DNA recombination of immunoglobulin (Ig) and T cell receptor (TCR) genes, which enables the recognition of highly diverse antigens and the elicitation of antigen-specific immune responses. This process is mediated by recombination-activating gene (Rag) 1 and Rag2 (Rag1/2), whose expression is strictly controlled in a cell type-specific manner; the expression of Rag1/2 genes represents a hallmark of lymphoid lineage commitment. Although Rag genes are known to be evolutionally conserved among jawed vertebrates, how Rag genes are regulated by lineage-specific transcription factors (TFs) and how their regulatory system evolved among vertebrates have not been fully elucidated. Here, we review the current body of knowledge concerning the cis-regulatory elements (CREs) of Rag genes and the evolution of the basic helix-loop-helix TF E protein regulating Rag gene CREs, as well as the evolution of the antagonist of this protein, the Id protein. This may help to understand how the adaptive immune system develops along with the evolution of responsible TFs and enhancers.
- adaptive immune system
- Rag1/2 gene enhancers
- E protein
- Id protein
- bHLH transcription factors
- T and B cell development
1. Introduction
Our body is protected from invading pathogens by immune responses, which are primarily mediated by two distinct types of cells, the adaptive and innate immune cells. These cells cooperatively function to induce inflammatory responses to eliminate pathogens from the body. Adaptive immune cells, such as T and B cells, elicit pathogen-specific immune responses through the recognition of specific antigens, while innate immune cells, including macrophages, neutrophils, dendritic cells, and histiocytes, are activated by pattern recognition receptors (PRRs), which recognize distinct microbial components. Because the Rag1/2 genes are exclusively expressed in T cell and B cell progenitor/precursor stages, their expression implies adaptive lymphoid lineage commitment [4].
As well as T cell, upon B cell lineage commitment from common lymphoid progenitors (CLPs) in the bone marrow, CLPs give rise to adaptive lymphoid cells (T and B cell), innate lymphoid cells (ILCs), and plasmacytoid dendritic cells (pDCs). Notably, ILCs and T cells show functional similarities in cytokine production, and they commonly express Bcl11b, Tcf1, Gata3, and Runx during their development and activation [17]. In T cells, an anti-silencer element (ASE), which is located 73 kb upstream of the Rag2 gene and is 8 kb in length, is essential for Rag1/2 gene expression in developing T cells, but not in developing B cells [20].
2. Regulation of Rag1/2 Gene by T or B Cell-Specific Enhancers
2.1. CREs for the Rag Gene, and Lineage-Specific Transcription Factors
The first wave of RAG expression is required for the recombination of TCRβ and Igh genes in pro-T and pro-B cells, respectively. After the β-selection of pro-T cells and pre-BCR selection of pro-B cells, Rag expression is transiently downregulated during the developmental transition toward precursor stages (DP and pre-B cells). Both in mouse and human, impairment or loss of Rag gene expression and functions results in severe combined immunodeficiency, resulting from developmental arrest at pro-T and pro-B cell stages [26,27]. Deletion of Erag, which is 23 kb upstream of the Rag2 gene, caused impaired Rag1/2 expression in pro-B cells and a moderate developmental block at the pro-B stage but did not affect the Rag gene expression in T cell development [22].
Tcf1, Bcl11b, Gata3, Runx1, Satb1, and Ikaros; B cells: Pax5, Ebf1, Foxo1, Ets1, Irf4, and Ikaros) [21,29,30,31]. These results indicated that E-protein binding to the T cell-specific Rag gene enhancer is required for T cell-specific spatial interactions to enhance Rag1/2 expression [14,21]. Notably, blocking E2A binding to the Rag1 gene promoter region (R1pro) by generating E-box motif mutations alone resulted in the complete loss of Rag1 expression without affecting Rag2 expression in both developing T and B cells, leading to developmental arrest at the pro-T and pro-B cell stages [21]. Taken together, these results strongly suggest that the activities of T cell-specific enhancer and Rag1 promoter depend on the binding of E2A to these regions and that E2A is a core TF that specifies the adaptive lymphoid cell identity through the regulation of Rag gene expression.
2.2. Evolution of Rag Gene Enhancer
Enhancer regions play a crucial role in precise pattern and amounts of gene expression during development, and divergence of the DNA sequence within enhancer region is considered to be related to the phenotypic variations among species [32]. This suggests that the phylogenetic conservation of DNA sequences within Rag gene enhancers reflect the evolution of Rag gene regulation. Thus, we investigated the conservation of R-TEn, R1B, and R2B regions and E-box motifs in these regions [21]. We found that DNA sequence similarities in R-TEn and R2B are readily observed among mammals, most birds, and reptiles; however, sequence similarities of these enhancers are not noticeable in the corresponding genomic regions of amphibians and fishes (Figure 2) [21]. Thus, we proposed that terrestrial animals evolutionarily acquired the E protein-mediated regulatory mechanisms as enhancers to increase the Rag gene expression, which induce higher expression of Rag genes and enable a diverse range of TCR and Ig gene recombination to protect our bodies from a wide range of pathogens.
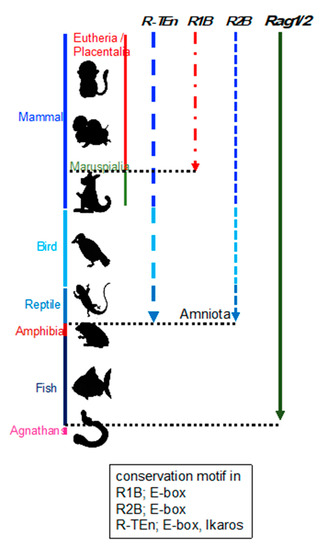
Figure 2. Schematic summary of the conservation of R-TEn, R1B, and R2B among vertebrates. Black, dotted lines indicate the border between placentaria and maruspialia, reptile and amphibia, and fish and agnathans. The conserved motifs in each enhancer region are shown in the box [21].
Regarding the evolution of AIS among vertebrates, cytidine deaminases CDA1 and CDA2 in jawless vertebrates are counterparts of Rag1 and Rag2 in jawed vertebrates and evolutionarily developed AIS as genome editors [33,34,35]. Furthermore, the recombination of Ig and TCR in fish seems to be more diverse than that in mammals, for example, the plasticity of T/B cells and the repertoire usage of TCR and Ig [36]. Given that the locations of B cell development among birds, reptiles, amphibians, and fish are different, it is reasonable that the variation in enhancer regions among species produces diversification of Rag1/2 gene regulation, such as timing. Considering this, it is surprising that both enhancer and promoter activities are critically controlled by E protein binding.
3. E Proteins and Id Proteins in Adaptive Lymphocyte Development
E proteins are basic helix-loop-helix (bHLH) transcription factors involved in multiple developmental processes. E proteins bind as homodimers or heterodimers to the E-box motif (CANNTG) within enhancer regions of their target genes. Id proteins contain an HLH domain missing the basic region that is essential for specific DNA binding and form heterodimers with bHLH proteins such as E proteins [39]. When the Id protein forms heterodimers with the E protein,
(Tcf3) is critically required for B cell lineage commitment [42,43] and the E2A gene encodes E12 and E47 proteins, which are generated by differential splicing [44,45]. In lymphoid progenitor cells, E2A orchestrates the B cell fate, along with Ebf1, Foxo1, and other TFs [12,46]. Upon T cell lineage commitment, E2A and HEB act in synergy to establish T cell identity and to suppress ILC development [18]. Likewise, HEB plays a role in iNKT cell development [47], and E2A and HEB also play important roles in the positive selection of DP thymocytes [48].
In B cell development, Id3 is induced in response to TGFβ signaling for survival during early B cell development [53]. Id3 is highly expressed in naïve mature B cells and downregulated in activated germinal center B (GCB) cells, while E2A protein abundance is low in naïve B cells but high in GCB cells to induce AID expression in cooperation with E2-2 [54,55]. In T cell development, Id3 is first upregulated by pre-TCR signaling in DN3 cells and further upregulated upon positive selection of TCR signaling in DP cells [56,57]. Furthermore, Id3 plays a key role in follicular helper T (TFH) and follicular cytotoxic T(TFC) cell development through the regulation of CXCR5 expression [60,61,62].
4. Evolution of E and Id Proteins
In this section, we address the question of how the E– Emc is a negative feedback regulator that prevents runaway self-stimulation of Da gene expression in Drosophila. Coupled transcriptional feedback loops maintain the widespread Emc expression that restrains Da activity to induce neurons [66], suggesting that the transcriptional regulation system by E and Id proteins is conserved from the common ancestor of mammals and Drosophila.
Three E protein homologs and two Id protein homologs were found in the lamprey (Petromyzon marinus) (Figure 3). A reconstructed maximum likelihood phylogenetic tree of E protein homologs indicates that homologs of jawed vertebrates form three clades for E2A, E2-2, and HEB. strongly suggest that these paralogs were generated through the widely recognized two rounds of whole genome duplication (WGD) in vertebrates [67]. It is plausible that ancestral jawed vertebrates probably had four paralogs for each of the E
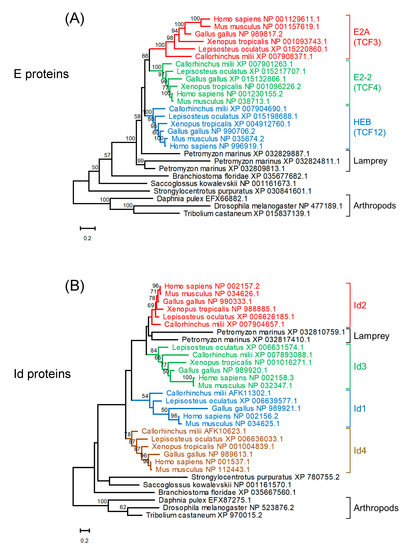
Figure 3. Maximum likelihood phylogenetic trees of homologs of E proteins (A) and Id proteins (B). Sequences were aligned using MAFFT (v7.453) [68] with default parameters. Tree reconstruction was performed using RAxML (version 8.2.12) [69] with the JTT + F substitution model and PROTGAMMA parameter with 100 bootstrap replicates. Phylogenetic trees were visualized using MEGA-X (version 10.2.4) [70]. Bootstrap values are given along the branches.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22115888
