Plants are constantly exposed to environmental stresses during their growth and development. Owing to their immobility, plants possess stress-sensing abilities and adaptive responses to cope with the abiotic and biotic stresses caused by extreme temperatures, drought, flooding, salinity, heavy metals and pathogens. Acyl-CoA-binding proteins (ACBPs), a family of conserved proteins among prokaryotes and eukaryotes, bind to a variety of acyl-CoA esters with different affinities and play a role in the transport and maintenance of subcellular acyl-CoA pools. In plants, studies have revealed ACBP functions in development and stress responses through their interactions with lipids and protein partners.
- abiotic stress
- acyl-CoA-binding proteins
- biotic stress
- lipids
- protein interactors
- stress signalling
1. Plant ACBPs
Plant ACBPs were first identified in Brassica napus L. (oilseed rape) as a 10-kDa homologue expressed in seeds, flowers and cotyledons [1]. It binds long-chain acyl-CoA esters [2], participates in acyl-CoA transport [3], maintains acyl-CoA pool [4], and regulates the activities of various enzymes including glycerol-3-phosphate acyltransferase [2], lysophosphatidylcholine acyltransferase [4] and lysophosphatidic acid acyltransferase [5]. The transport of acyl-CoA esters is important for the biosynthesis of lipids such as glycerolipids, ceramides and phospholipids, and studies have shown that the binding of phospholipids to ACBPs plays a role in plant growth and development as well as stress responses [6][7][8][9][10][11][12][13][14][15]. Following the discovery of BnACBP, similar 10-kDa ACBPs emerged in Arabidopsis thaliana [16], Gossypium hirsutum (cotton) [17], Ricinus communis (castor bean) [18], Digitalis lanata Ehrh. (Wolly Foxglove) [19], Oryza sativa (rice) [10], Vernicia fordii (tung tree) [20], Vitis vinifera (grape) [21], Helianthus annuus (sunflower) [22], Elaeis guineensis (oil palm) [23], Zea mays (maize) [24] and Glycine max (soybean) [25].
In plants such as Arabidopsis, rice, oilseed rape, oil palm, maize and soybean, ACBPs are classified into four main groups according to size and domains: Class I small ACBPs, Class II ACBPs containing ankyrin repeats, Class III large ACBPs and Class IV ACBPs containing kelch motifs (Table 1) [10][23][24][25][26]. Table 1 shows that Class I ACBPs range from 10 to 17 kDa, whereas the others comprised of a transmembrane domain, ankyrin repeats and/or kelch motifs, have molecular weights of 34 to 85 kDa [10][24]. Arabidopsis ACBPs are localised to the ER and plasma membrane (AtACBP1 and AtACBP2) [27][28], apoplast (AtACBP3) [29] and cytosol (AtACBP4 to AtACBP6) [8][30]. On the other hand, rice ACBPs are subcellularly localised to the cytosol (OsACBP1 to OsACBP3) [12], ER (OsACBP4) [12][31], apoplast (OsACBP5) [32] and peroxisomes (OsACBP6) [12]. In maize, transient expression of green fluorescent protein (GFP)-tagged Class I ZmACBP1 in Nicotiana benthamiana leaf epidermal cells revealed that ZmACBP1 was confined to the cytosol, Class II ZmACBP3 localised to the ER, whereas Class III and IV ZmACBP6 and ZmACBP7, respectively, were targeted to both the cytosol and the plasma membrane [24]. Oil palm Class II EgACBP2 contains an N-terminal transmembrane domain responsible for protein targeting to the plasma membrane, and two C-terminal ankyrin repeats which could mediate protein-protein interactions and other cellular activities [23]. Consistent with Protein Subcellular Localization Prediction Tool (PSORT) speculation, the sunflower Class I HaACBP6, which was transiently expressed in tobacco leaves, was localised to the cytosol and nucleus [22]. These results are summarised in Table 1.
Table 1. Characterization of plant ACBPs.
| Class | Protein Name | Signal Peptide | TM Domain | ACB Domain | Ankyrin Repeats | Kelch Motifs | Subcellular Locations | Size (kDa) |
|---|---|---|---|---|---|---|---|---|
| I | AtACBP6 | − | − | + | − | − | Cytosol | 10.4 |
| OsACBP1 | − | − | + | − | − | Cytosol | 10.2 | |
| OsACBP2 | − | − | + | − | − | Cytosol | 10.3 | |
| OsACBP3 | − | − | + | − | − | Cytosol | 17.7 | |
| ZmACBP1 | − | − | + | − | − | Cytosol | 10.1 | |
| HaACBP6 | − | − | + | − | − | Cytosol, Nucleus | 10.9 | |
| II | AtACBP1 | − | + | + | + | − | ER, PM | 37.5 |
| AtACBP2 | − | + | + | + | − | ER, PM | 38.5 | |
| OsACBP4 | + | + | + | + | − | ER | 36 | |
| ZmACBP3 | − | − | + | + | − | ER | 34.8 | |
| EgACBP2 | − | + | + | + | − | PM | ND | |
| III | AtACBP3 | + | + | + | − | − | Apoplast | 39.3 |
| OsACBP5 | + | + | + | − | − | ER | 61.2 | |
| ZmACBP6 | − | − | + | − | − | Cytosol, PM | 35.2 | |
| IV | AtACBP4 | − | − | + | − | + | Cytosol | 73.2 |
| AtACBP5 | − | − | + | − | + | Cytosol | 71 | |
| OsACBP6 | − | + | + | − | + | Peroxisomes | 71.4 | |
| ZmACBP7 | − | − | + | − | + | Cytosol, PM | 72.1 |
Abbreviations: ACB, acyl-CoA-binding; ACBP, acyl-CoA-binding protein; At, Arabidopsis thaliana; Eg, Elaeis guineensis; ER, endoplasmic reticulum; Ha, Helianthus annuus; kDa, kilodalton; ND, not determined; Os, Oryza sativa; PM, plasma membrane; TM, transmembrane; Zm, Zea mays; −, absent; +, present.
Using isothermal titration calorimetry (ITC), it has been reported that all recombinant ACBPs (rACBPs) bind acyl-CoA esters with varying affinities; rAtACBP1 and rAtACBP3 displayed high affinity to very-long-chain (VLC) species [33][29][34], while rAtACBP3 to rAtACBP6 and rOsACBPs to medium-chain species [10][35]. All rAtACBPs and rOsACBPs bind long-chain acyl-CoA esters at different affinities [36][10][15][29][37][38]. Moreover, rACBPs were shown to bind phospholipids, all Arabidopsis rAtACBPs bind PC [7][8][34][38][39], and rAtACBP1, rAtACBP2 and rAtACBP3 bind PA, lysoPC, and PE, respectively [6][9][34][40]. In contrast, all rice rOsACBPs bind PA and PC [10]. Besides binding with high affinity to 16:0-CoA, 18:0-CoA and 18:1-CoA, sunflower Class I rHaACBP6 and Class II rHaACBP1 also bind to several PC species [22][41]. In addition to phospholipid binding, Arabidopsis ACBPs were shown to interact with protein interactors (Table 2). AtACBPs interact with various transcription factors that activate the gene expression for downstream abscisic acid (ABA) or ethylene responses upon perception of stress stimuli [42][43][44]. These transcription factors include ABA-RESPONSIVE ELEMENT BINDING PROTEIN1 (AREB1) [44], RELATED TO APETALA2.12 (RAP2.12) [43] and ETHYLENE-RESPONSIVE ELEMENT BINDING PROTEIN (AtEBP) [42]. Furthermore, AtACBPs bind enzymes for sterol or phospholipid metabolisms such as PHOSPHOLIPASE Dα1 (PLDα1) [7], STEROL C4-METHYL OXIDASE1-1 (SMO1-1) [45], SMO1-2 [46] and LYSOPHOSPHOLIPASE2 (LYSOPL2) [6][47], which are important for membrane stability and repair as well as plant development. Thus far, only AtACBP2 interacts with FARNESYLATED PROTEIN6 (AtFP6) which may be involved in phospholipid repair following heavy metal-induced lipid peroxidation [36]. Lipid binding of ACBPs and their protein-protein interactions are now known to be important in regulating abiotic and biotic stress responses [48][36][6][7][44][8][9][14][49][50][51][52][53][54][55][56][57], as well as plant development including embryogenesis [46][39][45], seed dormancy [7], seed germination and development [7][44][58][59][60], cuticle development [61][33], pollen growth [62] and senescence [34][40].
Table 2. Abiotic and biotic stress responses mediated by plant ACBPs and/or their interactors.
| Proteins | Species | Acyl-CoA Binding | Phospholipid Binding | Protein Interactors | Stress Responses |
|---|---|---|---|---|---|
| AtACBP1 | A. thaliana | 16:0, 18:1, 18:2, 18:3, 20:4, 24:0, 25:0, 26:0 [33][29][63] | PC [7] PA [9] |
PLDα1 [7] | Freezing [9] |
| RAP2.12 [43][57][64] | Hypoxia [43][57] | ||||
| AREB1 [44] | Salinity, osmotic damage [44] | ||||
| − | Heavy metal [65] | ||||
| − | Pathogen [33] | ||||
| AtACBP2 | A. thaliana | 16:0, 18:1, 18:2, 18:3, 20:4 [36][29][37] | PC [39] lysoPC [6] |
AtEBP [42], RAP2.12 [43][57] | Hypoxia [42][43] |
| LYSOPL2 [6][47], AtFP6 [36] | Heavy metal [36][6][47] | ||||
| − | Drought [11] | ||||
| − | Salinity [15] | ||||
| − | Oxidation [36] | ||||
| AtACBP3 | A. thaliana | 12:0, 14:0, 16:0, 18:1, 18:2, 18:3, 20:4, 22:0, 24:0 [29][34][55][66] | PC [34] PE [34][40] |
− | Drought [61] |
| − | Hypoxia [66][67] | ||||
| − | Wounding [55] | ||||
| − | Pathogen [61][29][50] | ||||
| AtACBP4 | A. thaliana | 14:0, 16:0, 18:0, 18:1, 18:2, 18:3 [35][38] | PC [38] | AtEBP [48] | Pathogen [61][48] |
| − | Drought [61] | ||||
| − | Heavy metal [53] | ||||
| AtACBP6 | A. thaliana | 14:0, 16:0, 18:0, 18:1, 18:2, 18:3, 20:4 [16][35][38] | PC [8] | − | Freezing [8][52] |
| − | Drought [61] | ||||
| − | Wounding [54] | ||||
| − | Pathogen [61] | ||||
| OsACBP4 | O. sativa | 16:0, 18:0, 18:1, 18:2, 18:3 [10][15] | PC, PA [12] | − | Salinity [10][15] |
| OsACBP5 | O. sativa | 16:0, 18:3 [10][14] | PC, PA [12] | − | Pathogen [10][14][56] |
| − | Wounding [10] | ||||
| OsACBP6 | O. sativa | 18:1, 18:2 [10] | PC, PA [12] | − | Wounding [10] |
| ZmACBP1 | Z. mays | − | − | − | Salinity, drought [24] |
| ZmACBP3 | Z. mays | − | − | − | Salinity, drought [24] |
| ChACBP1 | Chlorella sp. | − | PC [13] | − | Freezing, salinity, oxidation, heavy metal [13] |
| VvACBP | V. vinifera | − | − | − | Freezing, heat, ER, pathogen [21] |
Abbreviations: AREB1, ABSCISIC ACID-RESPONSIVE ELEMENT BINDING PROTEIN1; AtEBP, Arabidopsis ETHYLENE-RESPONSIVE BINDING PROTEIN; AtFP6, Arabidopsis FARNESYLATED PROTEIN6; ER, endoplasmic reticulum; lysoPC, lysophosphatidylcholine; LYSOPL2, LYSOPHOSPHOLIPASE2; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PLDα1, PHOSPHOLIPASE Dα1; RAP2.12, RELATED TO APETALA2.12.
2. Membrane Lipids and ACBPs in Abiotic Stress Signalling
Plants are sessile, and therefore possess signalling and adaptive mechanisms to counteract abiotic and biotic stresses including cold, drought, salinity, oxidation, heavy metals, hypoxia and pathogen attack. Given the importance of plant ACBPs in development and stress responses, the roles of all Arabidopsis and rice ACBPs at different stages of plant growth were previously summarized by Du et al. [68] and are now updated (Table 2). Studies on the binding by ACBPs of acyl-CoA esters, membrane lipids and protein interactors have provided insights into the mechanistic events that occur when plants are exposed to various abiotic stresses (Figure 1).
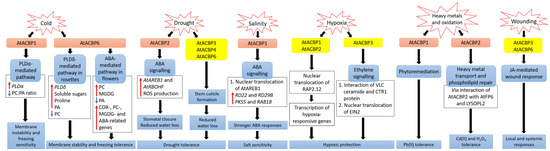
Figure 1. Signalling pathways associated with acyl-CoA-binding proteins (ACBPs) following abiotic stresses in Arabidopsis thaliana. In transgenic Arabidopsis Class II AtACBP1-overexpressors (OEs), PLDα1 was induced upon cold stress, causing a decrease in the ratio of PC to PA leading to membrane instability and freezing sensitivity [9]. In contrast, transgenic Arabidopsis Class I AtACBP6-OEs were conferred freezing tolerance via the PLDδ-mediated pathway in rosettes and the ABA-mediated pathway in flowers, resulting in changes in lipids, sugars and stress-related genes [8][52]. During drought, transgenic Arabidopsis Class II AtACBP2-OEs exhibited elevated AtAREB1 and AtRBOHF expression which led to ROS production, subsequent stomatal closure and reduced water loss [11]. Proper stem cuticle development conferred by Class I AtACBP6, Class III AtACBP3 or Class IV AtACBP4 protects wild-type Arabidopsis from water loss [61]. Under high salinity, AtACBP1 and AtAREB1 expression were upregulated in wild-type seeds [44]. The overexpression of AtACBP1 in transgenic Arabidopsis triggers nuclear translocation of AtAREB1, leading to the induction of stress marker genes (RD22 and RD29B) and AtAREB1 target genes (PKS5 and RAB18), thereby promoting stronger ABA responses during seed germination and seedling establishment [44]. When wild-type Arabidopsis undergoes hypoxia, the RAP2.12 transcription factor bound to AtACBP1 or AtACBP2, translocates to the nucleus and activates hypoxia-responsive gene transcription, conferring hypoxic protection [43][57][64][69]. Another hypoxic tolerance pathway involves the interaction of unsaturated VLC ceramide and the CTR1 protein with subsequent nuclear translocation of EIN2, resulting in the activation of CTR1-mediated ethylene signalling [67]. AtACBP1 is involved in phytoremediation and its overexpression in transgenic Arabidopsis confers Pb(II) tolerance [65]. AtACBP2 can interact with AtFP6 or LYSOPL2, mediating heavy metal transport and phospholipid repair respectively, and hence transgenic Arabidopsis AtACBP2-OEs were resistant to Cd(II) and Cd(II)-induced oxidative stress [36][6][47]. On wounding, the up-regulation of AtACBP3 and AtACBP6 expression in the wild type suggested their involvement in JA-mediated local and systemic wound responses [54][55]. Orange and yellow boxes indicate transgenic Arabidopsis AtACBP-OEs and wild-type Arabidopsis AtACBPs respectively, used in studies on abiotic stress. Blue boxes represent the signalling pathways. White boxes indicate the molecular events that occur along the signalling pathway. Red and blue arrows indicate increase and a decrease, respectively. Black arrows denote the flow of events. ABA, abscisic acid; ACBP, acyl-CoA-binding protein; AREB1, ABA-RESPONSIVE ELEMENT BINDING PROTEIN1; FP6, FARNESYLATED PROTEIN6; COR, COLD-RESPONSIVE; CTR1, CONSTITUTIVE TRIPLE RESPONSE1; EIN2, ETHYLENE-INSENSITIVE2; JA, jasmonic acid; LYSOPL2, LYSOPHOSPHOLIPASE2; MGDG, monogalactosyldiacylglycerol; PA, phosphatidic acid; PC, phosphatidylcholine; PKS5, PROTEIN KINASE SOS2-LIKE5; PLD, PHOSPHOLIPASE D; RAB18, RESPONSIVE TO ABA18; RAP2.12, RELATED TO APETALA2.12; RBOHF, RESPIRATORY BURST OXIDASE HOMOLOG F; RD, RESPONSIVE TO DESSICATION; ROS, reactive oxygen species; VLC, very-long-chain.
2.1. Cold Stress
AtACBP6-overexpressing (AtACBP6-OE) transgenic Arabidopsis rosettes and flowers are freezing tolerant (Figure 1) [8][52]. Northern-blot and Western-blot analyses showed that the expression of AtACBP6 and its protein in the wild type was induced at 48 h after 4°C cold treatment [8]. The atacbp6 mutant showed increased sensitivity to freezing temperature (−8°C) in contrast to the AtACBP6-OE plants [8]. Lipid profiles of rosettes upon freezing treatment of AtACBP6-OE transgenic Arabidopsis recorded decreases in PC and increases in PA, over the wild-type plants [8]. Furthermore, in vitro filter-binding assays revealed that rAtACBP6 binds PC, but not PA or lysoPC, suggesting a role for AtACBP6 in phospholipid metabolism in Arabidopsis [8]. On the other hand, in transgenic Arabidopsis AtACBP6-OE flowers, PC and monogalactosyldiacylglycerol (MGDG) levels were elevated while PA decreased [52]. In AtACBP6-OE rosettes, PHOSPHOLIPASE Dδ (PLDδ) was upregulated in the absence of COLD-RESPONSIVE (COR)-related gene induction [8], while flowers showed increased expression of COR-related genes and their transcription factors (C-repeat binding factors (CBFs), INDUCER OF CBF EXPRESSION1 (ICE1) and MYB15), PC-related genes, MGDG-related genes and ABA-related genes [52]. These results suggest a differential mechanism of freezing tolerance conferred by AtACBP6 in rosettes and flowers, possibly mediated by soluble sugar and proline accumulation and the ABA signalling pathway, respectively (Figure 1) [52].
Besides Class I AtACBP6, Arabidopsis Class II AtACBP1 also plays a role in freezing tolerance (Figure 1) [9]. AtACBP1-OE transgenic Arabidopsis plants were more cold-sensitive, accompanied by PC reduction and PA elevation, while atacbp1 plants were better protected from freezing arising from an increase in PC and a reduction in PA [9]. Although AtACBP1 and AtACBP6 belong to the same protein family, they play distinctive roles in cold tolerance. In vitro binding of rAtACBP1 to PA indicated possible enhanced PA interaction in AtACBP1-OE plants [7][9]. PLDα1, an important enzyme that catalyses the conversion of PC to PA, showed a higher gene expression in AtACBP1-OE plants than in atacbp1 [9]. In contrast, PLDδ expression decreased in the AtACBP1-OEs but increased in atacbp1 [9]. As AtACBP1 is localised to the ER and plasma membrane, it may maintain a membrane-associated PA pool through PA binding, thereby regulating the expression of PLDα1 and PLDδ [9].
Other than AtACBPs, grape VvACBP was upregulated in leaves upon cold and heat shock stresses in comparison to the nontreated control [21]. In maize, the expression levels of ZmACBP2, ZmACBP3, ZmACBP5 and ZmACBP6 were induced by cold stress while ZmACBP1, ZmACBP4, ZmACBP7, ZmACBP8 and ZmACBP9 mRNA levels declined after cold treatment [24]. These changes in expression levels depicted the potential roles of ZmACBPs in cold stress response which remain to be further elucidated. RNA-seq data analysis of the expression of soybean GmACBPs showed that only Class IV GmACBP11 was downregulated at 24 h after cold stress, whereas other GmACBPs displayed a lack of significant changes of expression in comparison to the nontreated control [25].
2.2. Drought Stress
Drought stress has received massive attention as it threatens worldwide crop production. ABA is a plant hormone that plays vital roles in many physiological processes including responses to abiotic stresses such as drought and salinity [70]. Under water deficiency, plants produce adaptive responses through the expression of various genes upon elevation of ABA [71][72]. Many signal transducers have been reported to participate in ABA signalling, including PA, diacylglycerol (DAG), phosphoinositides, reactive oxygen species (ROS), cyclic adenosine 5′-diphosphate ribose, sphingosine 1-phosphate and calcium [73][74][75][76][77][78][79][80][81][82].
Class II membrane-associated AtACBP2 responds to drought stress via ABA signalling (Figure 1) [11]. AtACBP2 expression was induced by ABA and drought treatment in wild-type Arabidopsis seedlings [11]. On top of that, transgenic Arabidopsis AtACBP2-OEs showed better drought tolerance than the wild type, whereas the atacbp2 mutant plants were more sensitive after drought treatment [11]. ABA-signalling genes including AREB1 and RESPIRATORY BURST OXIDASE HOMOLOG F (AtRBOHF) were upregulated in AtACBP2-OE before and after ABA treatment while AtRBOHD and ABA DEFICIENT2 (ABA2) increased only after ABA treatment. These results support the role of AtACBP2 in ABA signalling and hence in drought tolerance, as characterized by stomatal closure and reduced water loss [11].
It has been suggested that AtACBP3, AtACBP4 and AtACBP6 can regulate drought tolerance through stem cuticle formation (Figure 1) [61]. Transmission electron microscopy (TEM) showed that the leaves of the atacbp3, atacbp4 and atacbp6 mutants each had an abnormal and more permeable cuticle in comparison to the wild type, resulting in water loss after drought stress [61]. Furthermore, marked changes of cuticular wax and cutin monomer profiles in atacbp3, atacbp4 and atacbp6 single mutant plants depicted that AtACBPs play an important role in cuticle formation as well as in drought tolerance [61]. In soybean, expression profiles of roots were analysed by RNA-seq following dehydration stress [83]. Data mining of GmACBP expression by Azlan et al. [25] revealed that Class II (GmACBP3 and GmACBP4), Class III (GmACBP7) as well as Class IV (GmACBP9) were induced, suggesting that these GmACBPs play a role in drought response.
2.3. Salinity Stress
High salt in soil is detrimental to plant growth and development, and this in turn severely affects the crop yield worldwide. Salt stress can induce other stresses including osmotic stress, ionic stress and oxidative stress [84][85]. Osmotic stress arises from the reduction of water potential due to high amount of salt at the root surface, leading to a reduction in water uptake by the plant [86]. Ionic stress occurs as there is excessive uptake of sodium (Na+) and chloride (Cl+) ions by plant roots, which eventually accumulate in leaves [87]. Besides, ROS production also increases upon exposure to salt stress, causing oxidative stress in plants [88][89][90][91][92][93].
Salt sensing and signalling are complex. One of the early salt-signalling components are phospholipids, including polyphosphoinositides and PA [94][95]. PI signalling triggers the biosynthesis of phosphoinositides and JA-related proteins upon salt stress and can rapidly remodel soybean lipid composition for stress adaption [96]. Under salt stress, Na+ homeostasis is regulated by the SALT OVERLY SENSITIVE (SOS) pathway whereby Na+ influx promotes PLDα1 enzyme activity, causing a rise in PA levels [97]. Acting as a signal relay, PA activates MITOGEN-ACTIVATED PROTEIN KINASE6 (MPK6) which then phosphorylates SOS1, a potential intracellular Na+ sensor [98][99][100]. Several pld mutants exhibit enhanced sensitivity to salt stress [101].
ChACBP1, isolated from the algae (Chlorella sp.) JB6, was induced under various abiotic stresses including salinity, oxidation, heavy metals and cold stresses [13]. Given the binding of rChACBP1 protein to PC and the improved tolerance of yeast and Arabidopsis overexpressing ChACBP1 to abiotic stresses, these responses may be mediated through phospholipid metabolism [13]. Following NaCl or mannitol treatment of Arabidopsis seeds, the expression of AtACBP1 and its protein partner AtAREB1 were upregulated over the water-treated control [44]. The overexpression of AtACBP1 rendered higher sensitivity of transgenic Arabidopsis to NaCl or mannitol treatment during seed germination and seedling establishment over the wild type, whereas the atacbp1 mutant was less sensitive during seed germination but not seedling establishment (Figure 1) [44]. In transgenic Arabidopsis DsRed-AtAREB1/AtACBP1-OEs, the overexpression of AtACBP1 led to nuclear translocation of DsRed-AtAREB1 [44]. Salt and osmotic stress marker genes (RD22 and RD29B) and AtAREB1 target genes (PKS5 and RAB18) were also induced in AtACBP1-OEs [44]. These results suggested that enhanced AtAREB1 production in AtACBP1-OEs promotes stronger ABA responses during seed to seedling transition when AtAREB1 is released from AtACBP1 to enter the nucleus (Figure 1) [44]. A recent study revealed that the overexpression of OsACBP4 and AtACBP2 conferred salt resistance in both transgenic rice and Arabidopsis [15]. Four salinity-responsive elements in the OsACBP4 5′-flanking region were confirmed to interact with nuclear proteins from salt-treated rice [15]. On top of that, the up-regulation of genes encoding acyl-CoA synthase under salt stress and the binding of rOsACBP4 to long-chain acyl-CoA esters suggested that OsACBP4 may regulate salinity responses via lipid metabolism [15].
A recent study by Zhu et al. [24] showed that Class I ZmACBP1 and Class II ZmACBP3 gene expression was induced after NaCl or mannitol treatment. Transgenic Arabidopsis overexpressing ZmACBP1 and ZmACBP3 exhibited better growth and longer roots in comparison to the vector control [24]. The expression levels of the lipid metabolic genes (FAD2, DGAT, PLA2, PLC3, and ACX) and stress-responsive genes (COR47, AREB1, RAB, ABI1, RD29A, and RD29B) under NaCl or mannitol significantly increased in ZmACBP3-OEs compared to the wild type [24]. These results suggested that ZmACBP3 overexpression may enhance stress tolerance through changes in lipid metabolism which led to the induction of stress-responsive genes [24]. In soybean response to NaCl stress, in silico analysis of GmACBP expression from RNA-seq data exhibited induction of Class II GmACBP3, Class III GmACBP7 and Class IV GmACBP9, but decreases in Class I GmACBP2 and Class IV GmACBP10 [25]. As only Arabidopsis and rice Class II ACBPs have been reported in the NaCl response, the greater increase of Class III GmACBP7 than Class II GmACBP3 expression implied different roles for GmACBPs in soybean [25].
2.4. Hypoxic Stress
Plants need oxygen for respiration. Hypoxia happens when plants encounter oxygen deprivation, usually arising from flooding and soil waterlogging. Plants regulate their oxygen-sensing ability by transcription factors belonging to group VII of the ETHYLENE-RESPONSE FACTORS (ERF-VIIs) which are protected against proteasomal degradation only under hypoxia [43]. The stabilized ERF-VIIs can translocate to the nucleus and bind the HYPOXIA-RESPONSIVE PROMOTOR ELEMENT (HRPE) to drive the transcription of anaerobic genes [102]. ERF-VII transcription factor, AtEBP interacts with AtACBP2 via the ankyrin repeats although AtEBP is colocalised to the nucleus, whereas AtACBP2 is found on the plasma membrane [42]. Under aerobic conditions, RAP2.12 interacts with AtACBP1 and AtACBP2 at the plasma membrane, preventing its translocation to the nucleus and protecting it from N-end rule degradation [43]. When hypoxia arises, RAP2.12 is transported to the nucleus to activate the transcription of hypoxia-responsive genes (Figure 1) [69]. Polyunsaturated 18:3-CoA was proven to regulate the release of RAP2.12 from the plasma membrane upon hypoxia [57]. Upon submergence, wild-type Arabidopsis significantly accumulated polyunsaturated 18:3-CoA [57]. Confocal microscopy and immunoblot analysis showed that 18:3-CoA promoted stronger stabilization of RAP2.12-GFP, HYPOXIA RESPONSIVE ERF 1 (HRE1)-GFP and RAP2.3-GFP fusions [57]. In vitro pull-down assays revealed that both 18:0- and 18:3-CoAs suppress the interaction of AtACBP1 and ERF-VII, suggesting that 18:3-CoA can modulate the dissociation of the AtACBP1-ERF-VII complex when hypoxia arises [57]. Moreover, 18:3-CoA treatment of atacbp1 AtACBP2-RNAi lines indicates that AtACBP1 and AtACBP2 are important for the 18:3-CoA-induced stabilization of RAP2.12 and induction of hypoxia-responsive genes [57]. In addition, cellular energy depletion following hypoxia increased 18:1-CoA levels, triggering the dissociation of AtACBP1-bound RAP2.12 and its subsequent nuclear translocation for the activation of hypoxic gene transcription [64].
Other than Class II AtACBPs, AtACBP3 also plays a role in hypoxic response in Arabidopsis through binding of VLC acyl-CoA esters and regulation of fatty acid metabolism such as unsaturated VLC ceramides [66]. The interaction of unsaturated VLC ceramide with the CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) protein promoted nuclear translocation of ETHYLENE-INSENSITIVE2 (EIN2), triggering CTR1-mediated ethylene signalling for hypoxic protection in Arabidopsis (Figure 1) [67]. Besides ceramides and acyl-CoAs, other lipids including phospholipids, galactolipids, oxylipins, wax and cutin are important in plant hypoxic responses [103]. Upon submergence, total PC, PE and phosphatidylglycerol (PG) content declined but phosphatidylserine (PS), PA, PI, oxidized lipid, ceramide and hydroxyceramide levels increased significantly [66][67]. Moreover, significant increase of oxidized galactolipids [MGDG and digalactosyldiacylglycerol (DGDG)] and phospholipids (PC, PE and PG), arabidopsides and malondialdehyde (MDA), implied that an oxidative burst occurs during hypoxia or posthypoxic reoxygenation, leading to significant lipid peroxidation [57][67][104]. In addition, transcriptomic analyses have shown changes in the expression of genes encoding proteins essential for the ceramide and sphingolipid LCB biosynthesis [67], lipid transfer, and wax and cutin transport during submergence [105]. Moreover, JA biosynthesis genes were enhanced upon postsubmergence reoxygenation, implicating that oxylipins may modulate the posthypoxic reoxygenation response in plants [104].
2.5. Heavy Metal and Oxidative Stresses
Heavy metals such as lead [Pb(II)], cadmium [Cd(II)] and zinc [Zn(II)] are major pollutants threatening the environment and living organisms. Therefore, several studies have been performed to investigate the role of AtACBPs in response to heavy metal stresses [36][65]. Using metal-chelate affinity chromatography and fluorescence analysis using dansyl aziridine-labelled proteins, rAtACBP1 was reported to bind Pb(II) [65]. The overexpression of AtACBP1 in transgenic Arabidopsis showed better tolerance to Pb(II) stress, whereas the atacbp1 mutant was more sensitive to Pb(II) (Figure 1). Accumulation of Pb(II) in the shoots of AtACBP1-overexpressing plants suggested a possible role of AtACBP1 in Pb(II) phytoremediation [65]. Besides AtACBP1, the expression of AtACBP4 was also induced by Pb(II) in both Arabidopsis shoots and roots [53]. When transgenic Brassica juncea expressing AtACBP1 and AtACBP4 were grown in Pb(II)-containing media, Pb(II) accumulated in the cytosol of root tips and the vascular tissues, further corroborating to the function of AtACBPs in phytoremediation [53].
AtACBP2, on the other hand, is responsive to Cd(II). Although there was no accumulation of heavy metals in AtACBP2-overexpressing plants, the overexpression of AtACBP2 enhanced tolerance to Cd(II) and oxidative stress (hydrogen peroxide, H2O2) in transgenic Arabidopsis (Figure 1) [36]. In the plasma membrane, AtACBP2 interacts via its ankyrin repeats with AtFP6, which has a metal-binding motif [36]. In Arabidopsis roots, AtFP6 expression was induced after Cd(II) treatment [36]. The overexpression of AtFP6 conferred better Cd(II) resistance than the wild type, possibly by mediating heavy metal transport in plants [36]. LYSOPL2, another protein interactor of AtACBP2, is an intermediate of phospholipid metabolism and detoxifies lysoPC [6]. LYSOPL2 expression was induced by Zn(II) and H2O2 in Arabidopsis. The overexpression of LYSOPL2 in Arabidopsis exhibited enhanced tolerance to Cd(II) and H2O2 in comparison to the wild type, suggesting the involvement of LYSOPL2 in phospholipid repair following metal-induced lipid peroxidation (Figure 1) [6]. Possibly, the efficiency of membrane repair could be improved by the formation of an AtLYSOPL2-AtACBP2 complex, facilitated by lysoPC binding to AtACBP2 [47].
2.6. Wounding
In plants, wounding results following biotic attack (herbivores, insects and pathogens), mechanical damage or weather-induced damage, which may culminate in the entry of pathogens and nutrient loss. Mechanical injury triggers the transduction of mobile signals in the plants, leading to localised responses at the wound sites (local response) and distal responses in the undamaged tissues (systemic response) [106]. Cell wall-derived oligogalacturonides (OGs) and a polypeptide systemin are well-characterized wounding signals [107]. Upon wounding, systemin interacts with a cell-surface receptor to trigger several signalling events, including the release of linolenic acid from plant cell membranes and its conversion to 12 oxo-phytodienoic acid (OPDA) and JA [106][108][109]. The accumulation of JA in wounded plants subsequently activates various defence genes encoding proteinase inhibitor, thionin and enzymes involved in secondary metabolism [110].
Both Class I AtACBP6 and Class III AtACBP3 are involved in the local and systemic wound responses in Arabidopsis [54][55]. AtACBP6 and AtACBP3 proteins are localised to the companion cells, sieve elements and phloem [54][55]. On wounding, AtACBP6 and AtACBP3 were induced in Arabidopsis [54][55]. In comparison to atacbp3 and AtACBP3-RNAi plants, wound-responsive JA marker genes such as JASMONATE ZIM-DOMAIN10, VEGETATIVE STORAGE PROTEIN2 and LIPOXYGENASE2, were upregulated more significantly in locally wounded and systemic wild-type leaves [55]. Besides, lower levels of MeJA and oxylipin-related FAs, including C18:2-FA and C18:3-FA, were observed in atacbp3 and AtACBP3-RNAi over wild-type phloem exudates [55]. ITC data showed that rAtACBP3 binds medium and long-chain acyl-CoA esters but not MeJA, suggesting that AtACBP3 maintains FA pool but does not transport MeJA in the phloem [55]. Taken together, the evidence indicated that AtACBP3, a phloem-mobile protein, possibly regulates JA-mediated local and systemic wound responses by its binding to acyl-CoA esters (Figure 1). Besides AtACBP3 and AtACBP6, rice OsACBP5 and OsACBP6, as well as several maize ZmACBPs (ZmACBP1, ZmACBP2, ZmACBP5 and ZmACBP6), were rapidly induced after wound treatment [10][24]. However, their specific roles in wound response remain to be elucidated.
3. Membrane Lipids and ACBPs in Pathogen Defense
Under the natural environment, plants are always exposed to a variety of bacterial and fungal pathogens. Several AtACBPs, such as AtACBP1, AtACBP3, AtACBP4 and AtACBP6, and Class III OsACBP5, have been reported to participate in plant defence against infections caused by bacterial and fungal pathogens (Figure 2) [61][33][14][50][56]. AtACBP3 expression was induced in wild-type Arabidopsis following pathogen infection (Pseudomonas syringae pv tomato DC3000 and Botrytis cinerea) and treatments using pathogen elicitors (arachidonic acid) and defence-related phytohormones [1-aminocyclopropane-1-carboxylic acid (ACC), MeJA and salicylic acid (SA)] [50]. An S-box (TTTAA) regulatory element identified at the AtACBP3 5′-flanking region was verified by electrophoretic mobility shift assay (EMSA) to bind nuclear proteins from pathogen-infected Arabidopsis leaves [51]. In addition, overexpression of AtACBP3 led to constitutive activation of pathogenesis-related (PR) genes, including PR1, PR2 and PR5, H2O2 production and cell death (Figure 2) [50]. Following P. syringae pv tomato DC3000 infection, a lower bacterial count signified better protection of AtACBP3-OEs against the pathogen in comparison to the wild type and atacbp3 mutant [50]. To determine whether the upregulation of PR genes is associated with the NONEXPRESSOR OF PR-1 (NPR1) or CORONATINE-INSENSITIVE1 (COI1) signalling pathway, transgenic Arabidopsis of the AtACBP3-OEnpr1 line was subject to P. syringae treatment [50]. Results showed that the PR genes were downregulated in AtACBP3-OEnpr1 and they no longer exhibit enhanced resistance to P. syringae infection, implying that the pathogen protection of AtACBP3-OEs is mediated by the NPR1 signalling pathway [50]. As AtACBP3-OEs were more susceptible to necrotrophic fungus B. cinerea infection compared to atacbp3, AtACBP3 is believed to play a differential role in the plant defence response against necrotrophic and biotrophic pathogens [50]. Apart from abiotic stress, grape VvACBP which belongs to the same Class III as AtACBP3, also plays a role in pathogen defence (Figure 2) [21]. The expression of VvACBP in transgenic Arabidopsis conferred resistance to P. syringae pv tomato DC3000 and Colletotrichum higginsianum upon infection [21].
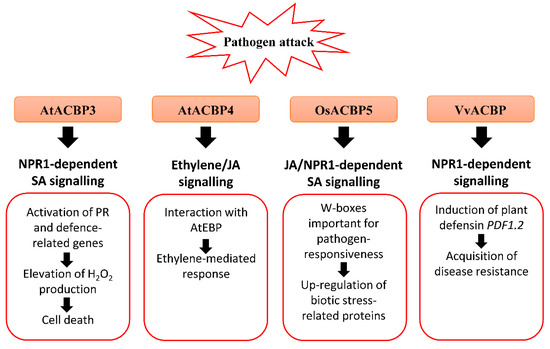
Figure 2. Biotic stress-related signalling pathways associated with acyl-CoA-binding proteins (ACBPs) in plants. Upon pathogen infection, the overexpression of Class III AtACBP3 in transgenic Arabidopsis thaliana led to constitutive activation of pathogenesis-related (PR) genes including PR1, PR2 and PR5, elevated H2O2 production and eventually cell death [50]. AtACBP3 plays a distinct role in the plant defence response against necrotrophic and biotrophic pathogens as transgenic Arabidopsis AtACBP3-overexpressors (OEs) were protected against the biotrophic pathogen (Pseudomonas syringae pv tomato DC3000) but not the necrotrophic pathogen (Botrytis cinerea) [50]. In wild-type Arabidopsis, the expression of Class IV AtACBP4 and AtEBP encoding a protein interactor of AtACBP4 were reported to be induced by B. cinerea infection, and ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) and methyl jasmonate (MeJA) treatments, suggesting that AtACBP4 and AtEBP are mediated by ethylene and/or JA signalling [48]. Rice Class III OsACBP5 protects transgenic Arabidopsis and rice plants against hemibiotrophs and biotrophs via NPR1-dependent SA signalling, and necrotrophs by JA signalling [14][56]. The OsACBP5 5′-flanking region contains W-boxes which were verified in pathogen-responsiveness of OsACBP5 [14]. Proteomic studies showed that eleven biotic stress-related proteins were upregulated by Rhizoctonia solani infection in transgenic Arabidopsis OsACBP5-OEs [56]. Grape Class III VvACBP conferred resistance to P. syringae and Colletotrichum higginsianum in transgenic Arabidopsis, possibly through the NPR1-mediated pathway following induction of PDF1.2, the gene encoding plant defensin [21]. Orange boxes represent the named ACBPs involved in the pathogen response. White boxes indicate the molecular events that occur along the signalling pathway. Black arrows denote the flow of signalling events. ACBP, acyl-CoA-binding protein; EBP, ETHYLENE-RESPONSIVE BINDING PROTEIN; JA, jasmonic acid; NPR1, NONEXPRESSOR OF PR-1; PR, pathogenesis-related; SA, salicylic acid.
The expression of Class IV AtACBP4 and its protein interactor AtEBP were elevated following B. cinerea infection, as well as the ethylene precursor ACC and MeJA treatments (Figure 2) [48]. The interaction of AtACBP4 and AtEBP, as confirmed by yeast two-hybrid and coimmunoprecipitation, suggests that plant pathogen defence may be mediated by ethylene and/or JA signalling [48]. Another study revealed that atacbp3, atacbp4 and atacbp6 single mutants exhibited a defective cuticle that resulted in compromised systemic acquired resistance (SAR) to fungal (B. cinerea and C. higginsianum) and bacterial (P. syringae) pathogens [61]. Furthermore, AtACBP1 which is also important for stem cuticle formation, is suggested to confer resistance to B. cinerea [33].
Apart from AtACBPs, recent studies have depicted that Class III OsACBP5, the homologue of AtACBP3, protects rice plants against representative necrotrophic (Rhizoctonia solani and Cercospora oryzae), hemibiotrophic (Magnaporthe oryzae and Fusarium graminearum) and biotrophic (Xanthomonas oryzae) phytopathogens (Figure 2) [14]. Transgenic rice OsACBP5-OEs demonstrated stronger disease resistance against all pathogens tested [14]. In addition, enhanced resistance of OsACBP5-OEs against hemibiotrophs and biotrophs is mediated by SA signalling, while that against the necrotrophic pathogen R. solani is regulated by JA signalling [14]. In the OsACBP5 5′-flanking region of the four W-boxes (pathogen-responsive cis-elements) identified, EMSAs showed that two of them bound nuclear proteins from wild-type rice infected with R. solani, C. oryzae, M. oryzae and X. oryzae [14]. Furthermore, transgenic rice expressing the construct of the OsACBP5 5′-flanking region containing both these W-boxes fused to the gene encoding β-glucuronidase (GUS) exhibited higher GUS activity upon SA, MeJA or R. solani treatment compared to the promoter deletion lacking both W-boxes [14]. These results suggest that the W-boxes are important in the pathogen-responsiveness of OsACBP5.
Both Lipidex assays and ITC showed that rOsACBP5 binds to 18:3-CoA esters, suggesting that 18:3-FA, a precursor for JA biosynthesis, plays a role in basal defence against fungal pathogens [10][14]. Furthermore, proteomic analysis revealed that eleven biotic stress-related proteins were upregulated by R. solani infection in transgenic Arabidopsis OsACBP5-OEs. These proteins include cell wall-related proteins such as FASCILIN-LIKE ARABINOGALACTAN-PROTEIN10 (FLA10), LEUCINE-RICH REPEAT EXTENSIN-LIKE PROTEINS (LRX4 and LRX5), XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE PROTEIN4 (XTH4) and PECTINESTERASE INHIBITOR18 (PME18), proteins involved in glucosinolate (GSL) degradation including GDSL-LIKE LIPASE23 (GLL23), EPITHIOSPECIFIER MODIFIER1 (ESM1), MYROSINASE1, MYROSINASE2, NITRILASE1 (NIT1), and a protein involved in JA synthesis, ALLENE OXIDE CYCLASE2 (AOC2), suggesting their potential in protection against R. solani [56].
Upon Phakopsora pachyrhizi fungal infection of soybean, seven GmACBPs comprising all four classes were detected in microarray data analysis [25]. The expression of Class I GmACBP2, Class II GmACBP4, Class III GmACBP5 and GmACBP6, and Class IV GmACBP11 declined after 6 h post inoculation (hpi) of soybean with avirulent Hawaii 94-1 and virulent Taiwan 80-2 strains [25]. In addition, Phytophthora sojae infection caused an induction of only Class IV GmACBP9 at 72 hpi, but a reduction of other GmACBPs such as Class I GmACBP2, Class III GmACBP5, GmACBP6 and GmACBP7 as well as Class IV GmACBP11 after 48 dpi [25]. Such differential changes in GmACBPs compared to other Class III ACBPs such as AtACBP3, VvACBP and OsACBP5 in pathogen response, suggested differential roles of GmACBPs in plant-pathogen interactions.
This entry is adapted from the peer-reviewed paper 10.3390/cells10051064
References
- Hills, M.J.; Dann, R.; Lydiate, D.; Sharpe, A. Molecular cloning of a cDNA from Brassica napus L. for a homologue of acyl-CoA-binding protein. Plant Mol. Biol. 1994, 25, 917–920.
- Brown, A.P.; Johnson, P.; Rawsthorne, S.; Hills, M.J. Expression and properties of acyl-CoA binding protein from Brassica napus. Plant Physiol. Biochem. 1998, 36, 629–635.
- Johnson, P.E.; Rawsthorne, S.; Hills, M.J. Export of acyl chains from plastids isolated from embryos of Brassica napus (L.). Planta 2002, 215, 515–517.
- Yurchenko, O.; Singer, S.D.; Nykiforuk, C.L.; Gidda, S.; Muller, R.T.; Moloney, M.M.; Weselake, R.J. Production of a Brassica napus low-molecular mass acyl-coenzyme A-binding protein in Arabidopsis alters the acyl-coenzyme A pool and acyl composition of oil in seeds. Plant Physiol. 2014, 165, 550–560.
- Brown, A.P.; Slabas, A.R.; Denton, H. Substrate selectivity of plant and microbial lysophosphatidic acid acyltransferases. Phytochemistry 2002, 61, 493–501.
- Gao, W.; Li, H.Y.; Xiao, S.; Chye, M.L. Acyl-CoA-binding protein 2 binds lysophospholipase 2 and lysoPC to promote tolerance to cadmium-induced oxidative stress in transgenic Arabidopsis. Plant J. 2010, 62, 989–1003.
- Du, Z.Y.; Chen, M.X.; Chen, Q.F.; Xiao, S.; Chye, M.L. Arabidopsis acyl-CoA-binding protein ACBP1 participates in the regulation of seed germination and seedling development. Plant J. 2013, 74, 294–309.
- Chen, Q.F.; Xiao, S.; Chye, M.L. Overexpression of the Arabidopsis 10-kilodalton acyl-coenzyme A-binding protein ACBP6 enhances freezing tolerance. Plant Physiol. 2008, 148, 304–315.
- Du, Z.Y.; Xiao, S.; Chen, Q.F.; Chye, M.L. Depletion of the membrane-associated acyl-coenzyme A-binding protein ACBP1 enhances the ability of cold acclimation in Arabidopsis. Plant Physiol. 2010, 152, 1585–1597.
- Meng, W.; Su, Y.C.F.; Saunders, R.M.K.; Chye, M.L. The rice acyl-CoA-binding protein gene family: phylogeny, expression and functional analysis. New Phytol. 2011, 189, 1170–1184.
- Du, Z.Y.; Chen, M.X.; Chen, Q.F.; Xiao, S.; Chye, M.L. Overexpression of Arabidopsis acyl-CoA-binding protein ACBP2 enhances drought tolerance. Plant Cell Environ. 2013, 36, 300–314.
- Meng, W.; Hsiao, A.S.; Gao, C.; Jiang, L.; Chye, M.L. Subcellular localization of rice acyl-CoA-binding proteins (ACBPs) indicates that OsACBP6::GFP is targeted to the peroxisomes. New Phytol. 2014, 203, 469–482.
- Qiao, K.; Wang, M.; Takano, T.; Liu, S. Overexpression of Acyl-CoA-Binding Protein 1 (ChACBP1) from saline-alkali-tolerant Chlorella sp. enhances stress tolerance in Arabidopsis. Front. Plant Sci. 2018, 9, 1772.
- Panthapulakkal Narayanan, S.; Lung, S.C.; Liao, P.; Lo, C.; Chye, M.L. The overexpression of OsACBP5 protects transgenic rice against necrotrophic, hemibiotrophic and biotrophic pathogens. Sci. Rep. 2020, 10, 14918.
- Guo, Z.H.; Pogancev, G.; Meng, W.; Du, Z.Y.; Liao, P.; Zhang, R.; Chye, M.L. The overexpression of rice ACYL-COA-BINDING PROTEIN4 improves salinity tolerance in transgenic rice. Environ. Exp. Bot. 2021, 183, 104349.
- Engeseth, N.J.; Pacovsky, R.S.; Newman, T.; Ohlrogge, J.B. Characterization of an acyl-CoA-binding protein from Arabidopsis thaliana. Arch. Biochem. Biophys. 1996, 331, 55–62.
- Reddy, A.S.; Ranganathan, B.; Haisler, R.M.; Swize, M.A. A cDNA encoding acyl-CoA-binding protein from cotton. Plant Physiol. 1996, 111, 348.
- Erber, A.; Horstmann, C.; Schobert, C. A cDNA clone for acyl-CoA-binding protein from castor bean. Plant Physiol. 1997, 114, 396.
- Metzner, M.; Ruecknagel, K.P.; Knudsen, J.; Kuellertz, G.; Mueller-Uri, F.; Diettrich, B. Isolation and characterization of two acyl-CoA-binding proteins from proembryogenic masses of Digitalis lanata Ehrh. Planta 2000, 210, 683–685.
- Pastor, S.; Sethumadhavan, K.; Ullah, A.H.J.; Gidda, S.; Cao, H.; Mason, C.; Chapital, D.; Scheffler, B.; Mullen, R.; Dyer, J.; et al. Molecular properties of the class III subfamily of acyl-coenzyme A binding proteins from tung tree (Vernicia fordii). Plant Sci. 2013, 203–204, 79–88.
- Takato, H.; Shimidzu, M.; Ashizawa, Y.; Takei, H.; Suzuki, S. An acyl-CoA-binding protein from grape that is induced through ER stress confers morphological changes and disease resistance in Arabidopsis. J. Plant Physiol. 2013, 170, 591–600.
- Aznar-Moreno, J.A.; Venegas-Calerón, M.; Du, Z.Y.; Garcés, R.; Tanner, J.A.; Chye, M.L.; Martínez-Force, E.; Salas, J.J. Characterization of a small acyl-CoA-binding protein (ACBP) from Helianthus annuus L. and its binding affinities. Plant Physiol. Biochem. 2016, 102, 141–150.
- Amiruddin, N.; Chan, P.L.; Azizi, N.; Morris, P.E.; Chan, K.L.; Ong, P.W.; Rosli, R.; Masura, S.S.; Murphy, D.J.; Sambanthamurthi, R.; et al. Characterization of oil palm acyl-CoA-binding proteins and correlation of their gene expression with oil synthesis. Plant Cell Physiol. 2019, 61, 735–747.
- Zhu, J.; Li, W.; Zhou, Y.; Pei, L.; Liu, J.; Xia, X.; Che, R.; Li, H. Molecular characterization, expression and functional analysis of acyl-CoA-binding protein gene family in maize (Zea mays). BMC Plant Biol. 2021, 21, 94.
- Azlan, N.S.; Guo, Z.H.; Yung, W.S.; Wang, Z.; Lam, H.M.; Lung, S.C.; Chye, M.L. In silico analysis of acyl-CoA-binding protein expression in soybean. Front. Plant Sci. 2021, 12, 646938.
- Raboanatahiry, N.H.; Yin, Y.; Chen, L.; Li, M. Genome-wide identification and phylogenic analysis of kelch motif containing ACBP in Brassica napus. BMC Genom. 2015, 16, 512.
- Chye, M.L.; Huang, B.Q.; Zee, S.Y. Isolation of a gene encoding Arabidopsis membrane associated acyl-CoA binding protein and immunolocalization of its gene product. Plant J. 1999, 18, 205–214.
- Li, H.Y.; Chye, M.L. Membrane localization of Arabidopsis acyl-CoA-binding protein ACBP2. Plant Mol. Biol. 2003, 51, 483–492.
- Leung, K.C.; Li, H.Y.; Xiao, S.; Tse, M.H.; Chye, M.L. Arabidopsis ACBP3 is an extracellularly targeted acyl-CoA-binding protein. Planta 2006, 223, 871–881.
- Xiao, S.; Li, H.Y.; Zhang, J.P.; Chan, S.W.; Chye, M.L. Arabidopsis acyl-CoA-binding proteins ACBP4 and ACBP5 are subcellularly localized to the cytosol and ACBP4 depletion affects membrane lipid composition. Plant Mol. Biol. 2008, 68, 571–583.
- Meng, W.; Chye, M.L. Rice acyl-CoA-binding proteins OsACBP4 and OsACBP5 are differentially localized in the endoplasmic reticulum of transgenic Arabidopsis. Plant Signal. Behav. 2014, 9, e29544.
- Liao, P.; Leung, K.P.; Lung, S.C.; Panthapulakkal Narayanan, S.; Jiang, L.; Chye, M.L. Subcellular localization of rice acyl-CoA-binding proteins ACBP4 and ACBP5 supports their non-redundant roles in lipid metabolism. Front. Plant Sci. 2020, 11, 331.
- Xue, Y.; Xiao, S.; Kim, J.; Lung, S.C.; Chen, L.; Tanner, J.A.; Suh, M.C.; Chye, M.L. Arabidopsis membrane-associated acyl-CoA-binding protein AtACBP1 is involved in stem cuticle formation. J. Exp. Bot. 2014, 18, 5473–5483.
- Xiao, S.; Gao, W.; Chen, Q.F.; Chan, S.W.; Zheng, S.X.; Ma, J.; Wang, M.; Welti, R.; Chye, M.L. Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell 2010, 22, 1463–1482.
- Leung, K.C.; Li, H.Y.; Mishra, G.; Chye, M.L. ACBP4 and ACBP5, novel Arabidopsis acyl-CoA-binding proteins with kelch motifs that bind oleoyl-CoA. Plant Mol. Biol. 2004, 55, 297–309.
- Gao, W.; Xiao, S.; Li, H.Y.; Tsao, S.W.; Chye, M.L. Arabidopsis thaliana acyl-CoA-binding protein ACBP2 interacts with a heavy-metal-binding farnesylated protein AtFP6. New Phytol. 2009, 181, 89–102.
- Chye, M.L.; Li, H.Y.; Yung, M.H. Single amino acid substitutions at the acyl-CoA-binding domain interrupt 14[C]palmitoyl-CoA binding of ACBP2, an Arabidopsis acyl-CoA-binding protein with ankyrin repeats. Plant Mol. Biol. 2000, 44, 711–721.
- Xiao, S.; Chen, Q.F.; Chye, M.L. Light-regulated Arabidopsis ACBP4 and ACBP5 encode cytosolic acyl-CoA-binding-proteins that bind phosphatidylcholine and oleoyl-CoA ester. Plant Physiol. Biochem. 2009, 47, 926–933.
- Chen, Q.F.; Xiao, S.; Qi, W.; Mishra, G.; Ma, J.; Wang, M.; Chye, M.L. The Arabidopsis acbp1acbp2 double mutant lacking acyl-CoA-binding proteins ACBP1 and ACBP2 is embryo lethal. New Phytol. 2010, 186, 843–855.
- Xiao, S.; Chye, M.L. The Arabidopsis thaliana ACBP3 regulates leaf senescence by modulating phospholipid metabolism and ATG8 stability. Autophagy 2010, 6, 802–804.
- Aznar-Moreno, J.A.; Venegas-Calerón, M.; Du, Z.Y.; Garcés, R.; Tanner, J.A.; Chye, M.L.; Martínez-Force, E.; Salas, J.J. Characterization and function of a sunflower (Helianthus annus L.) Class II acyl-CoA-binding protein. Plant Sci. 2020, 300, 110630.
- Li, H.Y.; Chye, M.L. Arabidopsis acyl-CoA-binding protein ACBP2 interacts with an ethylene-responsive element binding protein, AtEBP, via its ankyrin repeats. Plant Mol. Biol. 2004, 54, 233–243.
- Licausi, F.; Kosmacz, M.; Weits, D.A.; Guintoli, B.; Giorgi, F.M.; Voesenek, L.A.C.J.; Perata, P.; van Dongen, J.T. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 2011, 479, 419–422.
- Chen, M.X.; Hu, T.H.; Xue, Y.; Zhu, F.Y.; Du, Z.Y.; Lo, C.; Chye, M.L. Arabidopsis acyl-coenzyme-A-binding protein ACBP1 interacts with AREB1 and mediates salt and osmotic signaling in seed germination and seedling growth. Environ. Exp. Bot. 2018, 156, 130–140.
- Lung, S.C.; Liao, P.; Yeung, E.C.; Hsiao, A.S.; Xue, Y.; Chye, M.L. Acyl-CoA-binding protein ACBP1 modulates sterol synthesis during embryogenesis. Plant Physiol. 2017, 174, 1420–1435.
- Lung, S.C.; Liao, P.; Yeung, E.C.; Hsiao, A.S.; Xue, Y.; Chye, M.L. Arabidopsis ACYL-COA-BINDING PROTEIN1 interacts with STEROL C4-METHYL OXIDASE1-2 to modulate gene expression of homeodomain-leucine zipper IV transcription factors. New Phytol. 2018, 218, 183–200.
- Miao, R.; Lung, S.C.; Li, X.; Li, X.D.; Chye, M.L. Thermodynamic insights into an interaction between ACYL-CoA-BINDING PROTEIN2 and LYSOPHOSPHOLIPASE2 in Arabidopsis. J. Biol. Chem. 2019, 294, 6214–6226.
- Li, H.Y.; Xiao, S.; Chye, M.L. Ethylene- and pathogen-inducible Arabidopsis acyl-CoA-binding protein 4 interacts with an ethylene-responsive element binding protein. J. Exp. Bot. 2008, 59, 3997–4006.
- Xiao, S.; Chye, M.L. Arabidopsis ACBP1 overexpressors are Pb(II)-tolerant and accumulate Pb(II). Plant Signal. Behav. 2008, 3, 693–695.
- Xiao, S.; Chye, M.L. Overexpression of Arabidopsis acyl-CoA-binding protein 3 enhances NPR1-dependent plant resistance to Pseudomonas syringae pv. tomato DC3000. Plant Physiol. 2011, 156, 2069–2081.
- Zheng, S.X.; Xiao, S.; Chye, M.L. The gene encoding Arabidopsis acyl-CoA-binding protein 3 is pathogen-inducible and subject to circadian regulation. J. Exp. Bot. 2012, 63, 2985–3000.
- Liao, P.; Chen, Q.F.; Chye, M.L. Transgenic Arabidopsis flowers overexpressing acyl-CoA-binding protein ACBP6 are freezing tolerant. Plant Cell Physiol. 2014, 55, 1055–1071.
- Du, Z.Y.; Chen, M.X.; Chen, Q.F.; Gu, J.D.; Chye, M.L. Expression of Arabidopsis acyl-CoA binding proteins AtACBP1 and AtACBP4 confers Pb(II) accumulation in Brassica juncea roots. Plant Cell Environ. 2015, 38, 101–117.
- Ye, Z.W.; Lung, S.C.; Hu, T.H.; Chen, Q.F.; Suen, Y.L.; Wang, M.; Hoffmann-Benning, S.; Yeung, E.; Chye, M.L. Arabidopsis acyl-CoA-binding protein ACBP6 localizes in the phloem and affects jasmonate composition. Plant Mol. Biol. 2016, 92, 717–730.
- Hu, T.H.; Lung, S.C.; Ye, Z.W.; Chye, M.L. Depletion of Arabidopsis ACYL-COA-BINDING PROTEIN3 affects fatty acid composition in the phloem. Front. Plant Sci. 2018, 9, 2.
- Panthapulakkal Narayanan, S.; Liao, P.; Taylor, P.W.J.; Lo, C.; Chye, M.L. Overexpression of a monocot acyl-CoA-binding protein confers broad-spectrum pathogen protection in a dicot. Proteomics 2019, 19, 1800368.
- Zhou, Y.; Tan, W.J.; Xie, L.J.; Qi, H.; Yang, Y.C.; Huang, L.P.; Lai, Y.X.; Tan, Y.F.; Zhou, D.M.; Yu, L.J.; et al. Polyunsaturated linolenoyl-CoA modulates ERF-VII-mediated hypoxia signaling in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 330–348.
- Hsiao, A.S.; Haslam, R.P.; Michaelson, L.V.; Liao, P.; Chen, Q.F.; Sooriyaarachchi, S.; Mowbray, S.L.; Napier, J.A.; Tanner, J.A.; Chye, M.L. Arabidopsis cytosolic acyl-CoA-binding proteins ACBP4, ACBP5 and ACBP6 have overlapping but distinct roles in seed development. Biosci. Rep. 2015, 34, e00165.
- Guo, Z.H.; Haslam, R.P.; Michaelson, L.V.; Yeung, E.C.; Lung, S.C.; Napier, J.A.; Chye, M.L. The overexpression of rice ACYL-CoA-BINDING PROTEIN2 increases grain size and bran oil content in transgenic rice. Plant J. 2019, 100, 1132–1147.
- Guo, Z.H.; Ye, Z.W.; Haslam, R.P.; Michaelson, L.V.; Napier, J.A.; Chye, M.L. Arabidopsis cytosolic acyl-CoA binding proteins function in determining seed oil content. Plant Direct 2019, 3, e00182.
- Xia, Y.; Yu, K.; Gao, Q.M.; Wilson, E.V.; Navarre, D.; Kachroo, P.; Kachroo, A. Acyl CoA binding proteins are required for cuticle formation and plant responses to microbes. Front. Plant Sci. 2012, 3, 224.
- Hsiao, A.S.; Yeung, E.C.; Ye, Z.W.; Chye, M.L. The Arabidopsis cytosolic acyl-CoA-binding proteins play combinatory roles in pollen development. Plant Cell Physiol. 2015, 56, 322–333.
- Chye, M.L. Arabidopsis cDNA encoding a membrane-associated protein with an acyl-CoA binding domain. Plant Mol. Biol. 1998, 38, 827–838.
- Schmidt, R.R.; Fulda, M.; Paul, M.V.; Anders, M.; Plum, F.; Weits, D.A.; Kosmacz, M.; Larson, T.R.; Graham, I.A.; Beemster, G.T.S.; et al. Low-oxygen response is triggered by an ATP-dependent shift in oleoyl-CoA in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E12101–E12110.
- Xiao, S.; Gao, W.; Chen, Q.F.; Ramalingam, S.; Chye, M.L. Overexpression of membrane-associated acyl-CoA-binding protein ACBP1 enhances lead tolerance in Arabidopsis. Plant J. 2008, 54, 141–151.
- Xie, L.J.; Yu, L.J.; Chen, Q.F.; Wang, F.Z.; Huang, L.; Xia, F.N.; Zhu, T.R.; Wu, J.X.; Yin, J.; Liao, B.; et al. Arabidopsis acyl-CoA-binding protein ACBP3 participates in plant response to hypoxia by modulating very-long-chain fatty acid metabolism. Plant J. 2015, 81, 53–67.
- Xie, L.J.; Chen, Q.F.; Chen, M.X.; Yu, L.J.; Huang, L.; Chen, L.; Wang, F.Z.; Xia, F.N.; Zhu, T.R.; Wu, J.X.; et al. Unsaturation of very long-chain ceramides protects plant from hypoxia induced damages by modulating ethylene signaling in Arabidopsis. PLoS Genet. 2015, 11, e1005143.
- Du, Z.Y.; Arias, T.; Meng, W.; Chye, M.L. Plant acyl-CoA-binding proteins: an emerging family involved in plant development and stress responses. Prog. Lipid Res. 2016, 63, 165–181.
- Gibbs, D.J.; Lee, S.C.; Isa, N.M.; Gramuglia, S.; Fukao, T.; Bassel, G.W.; Correia, C.S.; Corbineau, F.; Theodolou, F.L.; Bailey-Serres, J.; et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 2011, 479, 415–418.
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012, 35, 53–60.
- Christmann, A.; Weiler, E.W.; Steudle, E.; Grill, E. A hydraulic signal in root-to-shoot signaling of water shortage. Plant J. 2007, 52, 167–174.
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signaling. Trends Plant Sci. 2010, 15, 395–401.
- Wu, Y.; Kuzma, J.; Maréchal, E.; Graeff, R.; Lee, H.C.; Foster, R.; Chua, N.H. Abscisic acid signaling through cyclic ADP-ribose in plants. Science 1997, 278, 2126–2130.
- Jacob, T.; Ritchie, S.; Assmann, S.M.; Gilroy, S. Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA 1999, 96, 12192–12197.
- Lemtiri-Chlieh, F.; MacRobbie, E.A.C.; Brearley, C.A. Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc. Natl. Acad. Sci. USA 2000, 97, 8687–8692.
- Pei, Z.M.; Murata, Y.; Benning, G.; Thomine, S.; Klüsener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signaling in guard cells. Nature 2000, 406, 731–734.
- Allen, G.J.; Chu, S.P.; Harrington, C.L.; Schumacher, K.; Hoffmann, T.; Tang, Y.Y.; Grill, E.; Schroeder, J.I. A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature 2001, 411, 1053–1057.
- Ng, C.K.; Carr, K.; McAinsh, M.R.; Powell, B.; Hetherington, A.M. Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature 2001, 410, 596–599.
- Scrase-Field, S.A.M.G.; Knight, M.R. Calcium: just a chemical switch? Curr. Opin. Plant Biol. 2003, 6, 500–506.
- Zhang, W.; Qin, C.; Zhao, J.; Wang, X. Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2004, 101, 9508–9513.
- Luan, S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009, 14, 37–42.
- Peters, C.; Li, M.; Narasimhan, R.; Roth, M.; Welti, R.; Wang, X. Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis. Plant Cell 2010, 22, 2642–2659.
- Belamkar, V.; Weeks, N.T.; Bharti, A.K.; Farmer, A.D.; Graham, M.A.; Cannon, S.B. Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genom. 2014, 15, 950.
- Ashraf, M. Some important physiological selection criteria for salt tolerance in plants. Flora 2004, 199, 361–376.
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539.
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499.
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681.
- Shen, B.; Jensen, R.G.; Bohnert, H.J. Increased resistance to oxidative stress in transgenic plants by targeting mannitol biosynthesis to chloroplasts. Plant Physiol. 1997, 113, 1177–1183.
- Tsugane, K.; Kobayashi, K.; Niwa, Y.; Ohba, Y.; Wada, K.; Kobayashi, H. A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell 1999, 11, 1195–1206.
- Hong, Z.; Lakkineni, K.; Zhang, Z.; Verma, D.P.S. Removal of feedback inhibition of delta(1)-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol. 2000, 122, 1129–1136.
- Zhu, J.; Fu, X.; Koo, Y.D.; Zhu, J.K.; Jenney, F.E.; Adams, M.W.; Zhu, Y.; Shi, H.; Yun, D.J.; Hasegawa, P.M.; et al. An enhancer mutant of Arabidopsis salt overly sensitive 3 mediates both ion homeostasis and the oxidative stress response. Mol. Cell. Biol. 2007, 27, 5214–5224.
- Hazman, M.; Hause, B.; Eiche, E.; Nick, P.; Riemann, M. Increased tolerance to salt stress in OPDA-deficient rice ALLENE OXIDE CYCLASE mutants is linked to an increased ROS-scavenging activity. J. Exp. Bot. 2015, 66, 3339–3352.
- Li, J.; Liu, J.; Wang, G.; Cha, J.Y.; Li, G.; Chen, S.; Li, Z.; Guo, J.; Zhang, C.; Yang, Y.; et al. A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 2015, 27, 908–925.
- Testerink, C.; Munnik, T. Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci. 2005, 10, 368–375.
- Testerink, C.; Munnik, T. Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 2011, 62, 2349–2361.
- Liu, A.; Xiao, Z.; Wang, Z.; Lam, H.M.; Chye, M.L. Galactolipid and phospholipid profile and proteome alterations in soybean leaves at the onset of salt stress. Front. Plant Sci. 2021, 12, 644408.
- Yu, L.; Nie, J.; Cao, C.; Jin, Y.; Yan, M.; Wang, F.; Liu, J.; Xiao, Y.; Liang, Y.; Zhang, W. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 2010, 188, 762–773.
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 2002, 14, 465–477.
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441.
- Shabala, L.; Cuin, T.A.; Newman, I.A.; Shabala, S. Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants. Planta 2005, 222, 1041–1050.
- Bargmann, B.O.R.; Laxalt, A.M.; ter Riet, B.; van Schooten, B.; Merquiol, E.; Testerink, C.; Haring, M.A.; Bartels, D.; Munnik, T. Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol. 2009, 50, 78–89.
- Gasch, P.; Fundinger, M.; Müller, J.T.; Lee, T.; Bailey-Serres, J.; Mustropha, A. Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 2016, 28, 160–180.
- Xie, L.J.; Zhou, Y.; Chen, Q.F.; Xiao, S. New insights into the role of lipids in plant hypoxia responses. Prog. Lipid Res. 2021, 81, 101072.
- Yuan, L.B.; Dai, Y.S.; Xie, L.J.; Yu, L.J.; Zhou, Y.; Lai, Y.X.; Yang, Y.C.; Xu, L.; Chen, Q.F.; Xiao, S. Jasmonate regulates plant responses to postsubmergence reoxygenation through transcriptional activation of antioxidant synthesis. Plant Physiol. 2017, 173, 1864–1880.
- Kim, H.; Choi, D.; Suh, M.C. Cuticle ultrastructure, cuticular lipid composition, and gene expression in hypoxia-stressed Arabidopsis stems and leaves. Plant Cell Rep. 2017, 36, 815–827.
- Farmer, E.E.; Ryan, C.A. Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell 1992, 4, 129–134.
- Roberts, K. Potential awareness of plants. Nature 1992, 360, 14–15.
- Meindl, T.; Boller, T.; Felix, G. The plant wound hormone systemin binds with the N-terminal part to its receptor but needs the C-terminal part to activate it. Plant Cell 1998, 10, 1561–1570.
- Scheer, J.M.; Ryan, C.A. A 160-kD Systemin receptor on the surface of Lycopersicon peruvianum suspension-cultured cells. Plant Cell 1999, 11, 1525–1535.
- Creelman, R.A.; Mullet, J.E. Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. 1997, 48, 355–381.
