Chronic infection by the hepatitis C virus (HCV) is a major cause of liver diseases, predisposing to fibrosis and hepatocellular carcinoma. Liver fibrosis is characterized by an overly abundant accumulation of components of the hepatic extracellular matrix, such as collagen and elastin, with consequences on the properties of this microenvironment and cancer initiation and growth. This review will provide an update on mechanistic concepts of HCV-related liver fibrosis/cirrhosis and early stages of carcinogenesis, with a dissection of the molecular details of the cross-talk during disease progression between hepatocytes, the extracellular matrix and hepatic stellate cells.
- liver fibrosis
- cirrhosis
- chronic hepatitis C
- carcinogenesis
- extracellular matrix
1. Introduction
|
Virus |
HBV |
HCV |
|---|---|---|
|
Viral family |
Hepadnaviridae |
Flaviviridae |
|
Genome |
DNA and cccDNA |
RNA |
|
Life cycle |
Genome integration, expression of HBx protein, insertional activation of cellular oncogenes, cccDNA (minichromosome) |
Exclusively cytoplasmic |
|
Persistence |
Nucleus-located cccDNA |
Chronic inflammation, oxidative stress, alterations in cellular signaling and metabolism |
2. Main Actors of Liver Fibrosis
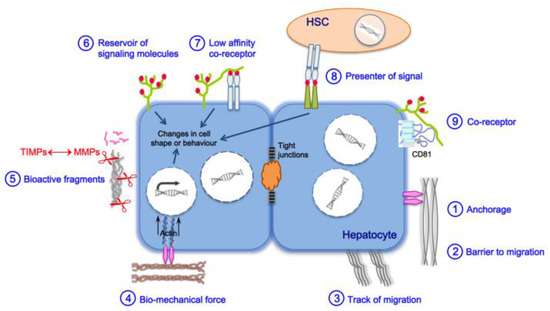
3. Liver Fibrosis and Cirrhosis
3.1. General Pan-Etiology Features
3.2. Features Linked to HCV Pathogenesis
|
HCV Proteins |
ECM Proteins or Cytokines |
|---|---|
|
Capsid core |
LOX ∞ [61] Procollagen I ∞ [62] Collagen I ∞ [61] MMP-2 ∞ [58] |
|
MMP-9 ∞ [63] |
|
|
COX-2 ∞ [63] |
|
|
Syndecan-1 * [31] |
|
|
Thrombospondin-1 ∞ [61] |
|
|
CTGF ∞ [58] |
|
|
TGF-β2 ◊ [67] |
|
|
Endoglin ∞ [68] |
|
|
Envelope glycoproteins E1 and/or E2 |
Glypican-3 * [69] |
|
TGF-β1 ◊ [66] |
|
|
Cysteine autoprotease NS2 |
MICA ∞ [70] TGF-β2 ◊ [67] |
|
Serine protease and helicase NS3 |
Procollagen I ∞ [62] MMP-9 ∞ [71] |
|
COX-2 ∞ [71] |
|
|
Thrombospondin-1 [72] |
|
|
Osteopontin * [64] |
|
|
TGF-β type I receptor * [73] |
|
|
NS3 with its cofactor NS4A |
MMP-9 ∞ [71] |
|
COX-2 ∞ [71] MICA ∞ [74] |
|
|
NS4B |
MMP-2 ∞ [76] |
|
NS5A |
MMP-2 ∞ [63] |
|
MMP-9 ∞ [63] |
|
|
COX-2 ∞ [63] |
|
|
Thrombospondin-1 ∞ [72] |
|
|
Osteopontin * [64] |
|
|
RNA-dependent RNA polymerase NS5B |
Osteopontin * [64] |
|
MICA ∞ [70] TGF-β ◊ [75] |
|
ECM Proteins/Cytokine |
F0/F1 |
F2 |
F3 |
F4 |
HCC |
References |
|---|---|---|---|---|---|---|
|
Collagens I, III, V |
F1 |
|||||
|
Collagen XII |
||||||
|
Collagen XIV |
||||||
|
Collagen XVI |
[59] |
|||||
|
Collagen XVIII |
[59] |
|||||
|
PIIINP |
F1 |
|||||
|
MMP-2, -7, -9 |
F1 |
|||||
|
TIMP-1 |
||||||
|
ADAM-TS1 |
[93] |
|||||
|
ADAM-TS2 |
[94] |
|||||
|
Xylosyltransferase-2 |
F1 |
|||||
|
Glypican-3 |
||||||
|
Hyaluronic acid |
||||||
|
Decorin |
F1 |
[92] |
||||
|
Biglycan |
[59] |
|||||
|
Fibromodulin |
[60] |
|||||
|
Lumican |
||||||
|
Versican |
F1 |
|||||
|
Tenascin-C |
||||||
|
Osteopontin |
F1 |
|||||
|
Fibronectin |
||||||
|
Fibronectin isoforms |
[108] |
|||||
|
Elastin |
||||||
|
MFAP-4 † |
F1 |
|||||
|
Fibulin-5 |
[84] |
|||||
|
TGF-β1 (protein, mRNA) |
||||||
|
TGF-β1 (serum levels) |
F1 |
|||||
|
TGF-β2 |
F1 |
F0 |
[67] |
|||
|
Endoglin (protein, serum levels) |
[100] |
|||||
|
Endoglin (mRNA) § |
[68] |
a Color codes: green, upregulation; dark green: higher upregulation; blue, downregulation; dark blue: higher downregulation; grey, no change; magenta, no correlation with liver fibrosis stage. † MFAP-4, microfibrillar-associated protein-4 (associated with elastin fibers). § Endoglin mRNA was found upregulated in chronically HCV-infected patients compared to noninfected patients but not correlating with liver fibrosis stage.
4. Are HSCs Direct Targets of HCV Infection?
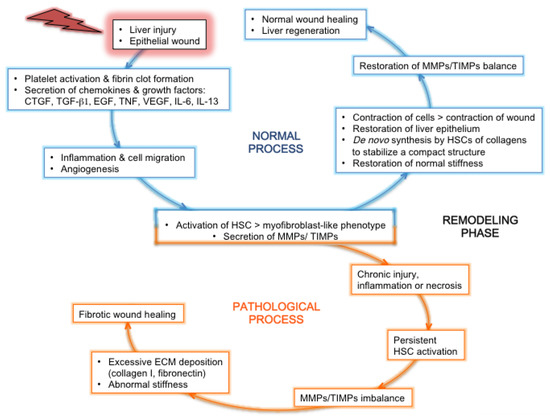
5. Fibrosis Reversal in the Era of DAAs in HCV-Induced Liver Fibrosis
In the era of DAAs which raise hope of eradicating HCV, HCV infection still remains a leading cause of hepatic failure due to advanced liver disease and HCC, because curing the infection does not fully restore liver homeostasis. Furthermore, DAA treatment alone may not be sufficient for a complete cure of fibrosis, as several factors other than the virus contribute to liver deterioration. Lastly, patients under antiviral therapy variably respond to the regression of fibrosis. The mechanism of HCV-induced liver disease is a multifaceted process, since various host genes are altered, and since host cells respond to infection/viral components by mobilizing or producing enzymes, growth factors and chemokines which activate quiescent HSCs. HCV chronic infection leads to a deep remodeling of the entire liver ECM architecture, through direct interactions between viral, ECM and cellular proteins, and through indirect effects (e.g. promotion of oxidative and ER stress, of inflammation, of stemness). HCV-induced overexpression of TGF-b, the most potent profibrogenic cytokine, contributes to HCV replication, and to the activation of HSCs, the promotion of their survival, and the inhibition of HSC apoptosis, mechanisms by which liver disease progresses. Consequently, several general mechanisms involved in liver fibrosis/cirrhosis development contribute to tumorigenesis. TGF-b signaling facilitates HCV replication in hepatocytes, and could promote survival of pre-cancerous cells; furthermore, HCV replicates at higher rates in liver cancer stem cells.
Thus, efforts toward a deeper comprehension of host/virus/ECM interactions, and of the underlying mechanisms by which hepatic dysfunctions emerge, spread and persist after HCV infection are therefore still needed, in order to develop therapies that cure liver disease in addition to curing infection.References.
- Plummer, M.; de Martel, C.; Vignat, J.; Ferlay, J.; Bray, F.; Franceschi, S. Global Burden of Cancers Attributable to Infections in 2012: A Synthetic Analysis. Lancet Glob Health 2016, 4, e609-616, doi:10.1016/S2214-109X(16)30143-7.
- Villanueva, A. Hepatocellular Carcinoma. N Engl J Med 2019, 380, 1450–1462, doi:10.1056/NEJMra1713263.
- Mitchell, J.K.; Lemon, S.M.; McGivern, D.R. How Do Persistent Infections with Hepatitis C Virus Cause Liver Cancer? Curr Opin Virol 2015, 14, 101–108, doi:10.1016/j.coviro.2015.09.003.
- Lemon, S.M.; McGivern, D.R. Is Hepatitis C Virus Carcinogenic? Gastroenterology 2012, 142, 1274–1278, doi:10.1053/j.gastro.2012.01.045.
- WHO Combating Hepatitis B and C to Reach Elimination by 2030. Advocacy Brief 2016.
- Polaris Observatory HCV Collaborators Global Prevalence and Genotype Distribution of Hepatitis C Virus Infection in 2015: A Modelling Study. Lancet Gastroenterol Hepatol 2017, 2, 161–176, doi:10.1016/S2468-1253(16)30181-9.
- WHO | Hepatitis C Available online: http://www.who.int.gate2.inist.fr/mediacentre/factsheets/fs164/en/ (accessed on 29 January 2018).
- Hajarizadeh, B.; Grebely, J.; Dore, G.J. Epidemiology and Natural History of HCV Infection. Nat Rev Gastroenterol Hepatol 2013, 10, 553–562, doi:10.1038/nrgastro.2013.107.
- Nault, J.-C.; Colombo, M. Hepatocellular Carcinoma and Direct Acting Antiviral Treatments: Controversy after the Revolution. J. Hepatol. 2016, 65, 663–665, doi:10.1016/j.jhep.2016.07.004.
- Baumert, T.F.; Jühling, F.; Ono, A.; Hoshida, Y. Hepatitis C-Related Hepatocellular Carcinoma in the Era of New Generation Antivirals. BMC Med 2017, 15, 52, doi:10.1186/s12916-017-0815-7.
- Hamdane, N.; Jühling, F.; Crouchet, E.; El Saghire, H.; Thumann, C.; Oudot, M.A.; Bandiera, S.; Saviano, A.; Ponsolles, C.; Roca Suarez, A.A.; et al. HCV-Induced Epigenetic Changes Associated With Liver Cancer Risk Persist After Sustained Virologic Response. Gastroenterology 2019, 156, 2313-2329.e7, doi:10.1053/j.gastro.2019.02.038.
- Hengst, J.; Falk, C.S.; Schlaphoff, V.; Deterding, K.; Manns, M.P.; Cornberg, M.; Wedemeyer, H. Direct-Acting Antiviral-Induced Hepatitis C Virus Clearance Does Not Completely Restore the Altered Cytokine and Chemokine Milieu in Patients With Chronic Hepatitis C. J. Infect. Dis. 2016, 214, 1965–1974, doi:10.1093/infdis/jiw457.
- Akuta, N.; Suzuki, F.; Hirakawa, M.; Kawamura, Y.; Sezaki, H.; Suzuki, Y.; Hosaka, T.; Kobayashi, M.; Kobayashi, M.; Saitoh, S.; et al. Amino Acid Substitutions in Hepatitis C Virus Core Region Predict Hepatocarcinogenesis Following Eradication of HCV RNA by Antiviral Therapy. J. Med. Virol. 2011, 83, 1016–1022, doi:10.1002/jmv.22094.
- Takeda, H.; Takai, A.; Iguchi, E.; Mishima, M.; Arasawa, S.; Kumagai, K.; Eso, Y.; Shimizu, T.; Takahashi, K.; Ueda, Y.; et al. Oncogenic Transcriptomic Profile Is Sustained in the Liver after the Eradication of the Hepatitis C Virus. Carcinogenesis 2021, doi:10.1093/carcin/bgab014.
- Paul, D.; Madan, V.; Bartenschlager, R. Hepatitis C Virus RNA Replication and Assembly: Living on the Fat of the Land. Cell Host Microbe 2014, 16, 569–579, doi:10.1016/j.chom.2014.10.008.
- El Sebae, G.K.; Malatos, J.M.; Cone, M.-K.E.; Rhee, S.; Angelo, J.R.; Mager, J.; Tremblay, K.D. Single-Cell Murine Genetic Fate Mapping Reveals Bipotential Hepatoblasts and Novel Multi-Organ Endoderm Progenitors. Development 2018, 145, doi:10.1242/dev.168658.
- Lee, Y.A.; Wallace, M.C.; Friedman, S.L. Pathobiology of Liver Fibrosis: A Translational Success Story. Gut 2015, 64, 830–841, doi:10.1136/gutjnl-2014-306842.
- Rauterberg, J.; Voss, B.; Pott, G.; Gerlach, U. Connective Tissue Components of the Normal and Fibrotic Liver. I. Structure, Local Distribution and Metabolism of Connective Tissue Components in the Normal Liver and Changes in Chronic Liver Diseases. Klin. Wochenschr. 1981, 59, 767–779.
- Friedman, S.L. Hepatic Stellate Cells: Protean, Multifunctional, and Enigmatic Cells of the Liver. Physiol. Rev. 2008, 88, 125–172, doi:10.1152/physrev.00013.2007.
- De Minicis, S.; Seki, E.; Uchinami, H.; Kluwe, J.; Zhang, Y.; Brenner, D.A.; Schwabe, R.F. Gene Expression Profiles during Hepatic Stellate Cell Activation in Culture and in Vivo. Gastroenterology 2007, 132, 1937–1946, doi:10.1053/j.gastro.2007.02.033.
- Lin, X.Z.; Horng, M.H.; Sun, Y.N.; Shiesh, S.C.; Chow, N.H.; Guo, X.Z. Computer Morphometry for Quantitative Measurement of Liver Fibrosis: Comparison with Knodell’s Score, Colorimetry and Conventional Description Reports. J. Gastroenterol. Hepatol. 1998, 13, 75–80.
- Duarte, S.; Baber, J.; Fujii, T.; Coito, A.J. Matrix Metalloproteinases in Liver Injury, Repair and Fibrosis. Matrix Biology: Journal of the International Society for Matrix Biology 2015, 44–46, 147–156, doi:10.1016/j.matbio.2015.01.004.
- Rojkind, M.; Giambrone, M.A.; Biempica, L. Collagen Types in Normal and Cirrhotic Liver. Gastroenterology 1979, 76, 710–719.
- Kagan, H.M. Lysyl Oxidase: Mechanism, Regulation and Relationship to Liver Fibrosis. Pathol. Res. Pract. 1994, 190, 910–919, doi:10.1016/S0344-0338(11)80995-7.
- Martinez-Hernandez, A.; Amenta, P.S. The Extracellular Matrix in Hepatic Regeneration. FASEB J. 1995, 9, 1401–1410.
- Iozzo, R.V.; Schaefer, L. Proteoglycan Form and Function: A Comprehensive Nomenclature of Proteoglycans. Matrix Biol. 2015, 42, 11–55, doi:10.1016/j.matbio.2015.02.003.
- Kanta, J. Elastin in the Liver. Front Physiol 2016, 7, 491, doi:10.3389/fphys.2016.00491.
- Musso, O.; Rehn, M.; Saarela, J.; Théret, N.; Liétard, J.; Hintikka, null; Lotrian, D.; Campion, J.P.; Pihlajaniemi, T.; Clément, B. Collagen XVIII Is Localized in Sinusoids and Basement Membrane Zones and Expressed by Hepatocytes and Activated Stellate Cells in Fibrotic Human Liver. Hepatology 1998, 28, 98–107, doi:10.1002/hep.510280115.
- Ricard-Blum, S.; Vallet, S.D. Fragments Generated upon Extracellular Matrix Remodeling: Biological Regulators and Potential Drugs. Matrix Biol. 2019, 75–76, 170–189, doi:10.1016/j.matbio.2017.11.005.
- Sun, S.; Song, Z.; Cotler, S.J.; Cho, M. Biomechanics and Functionality of Hepatocytes in Liver Cirrhosis. J Biomech 2014, 47, 2205–2210, doi:10.1016/j.jbiomech.2013.10.050.
- Grigorov, B.; Reungoat, E.; Gentil Dit Maurin, A.; Varbanov, M.; Blaising, J.; Michelet, M.; Manuel, R.; Parent, R.; Bartosch, B.; Zoulim, F.; et al. Hepatitis C Virus Infection Propagates through Interactions between Syndecan-1 and CD81, and Impacts the Hepatocyte Glycocalyx. Cell. Microbiol. 2017, 19, doi:10.1111/cmi.12711.
- Wells, J.M.; Gaggar, A.; Blalock, J.E. MMP Generated Matrikines. Matrix Biol. 2015, 44–46, 122–129, doi:10.1016/j.matbio.2015.01.016.
- Schuppan, D.; Ashfaq-Khan, M.; Yang, A.T.; Kim, Y.O. Liver Fibrosis: Direct Antifibrotic Agents and Targeted Therapies. Matrix Biol. 2018, 68–69, 435–451, doi:10.1016/j.matbio.2018.04.006.
- Ricard-Blum, S.; Baffet, G.; Théret, N. Molecular and Tissue Alterations of Collagens in Fibrosis. Matrix Biol. 2018, doi:10.1016/j.matbio.2018.02.004.
- Jung, Y.; Witek, R.P.; Syn, W.-K.; Choi, S.S.; Omenetti, A.; Premont, R.; Guy, C.D.; Diehl, A.M. Signals from Dying Hepatocytes Trigger Growth of Liver Progenitors. Gut 2010, 59, 655–665, doi:10.1136/gut.2009.204354.
- Wynn, T.A. Cellular and Molecular Mechanisms of Fibrosis. J Pathol 2008, 214, 199–210, doi:10.1002/path.2277.
- Tsochatzis, E.A.; Bosch, J.; Burroughs, A.K. Liver Cirrhosis. Lancet 2014, 383, 1749–1761, doi:10.1016/S0140-6736(14)60121-5.
- Zhao, S.-X.; Li, W.-C.; Fu, N.; Kong, L.-B.; Zhang, Q.-S.; Han, F.; Ren, W.-G.; Cui, P.; Du, J.-H.; Wang, B.-Y.; et al. CD14+ Monocytes and CD163+ Macrophages Correlate with the Severity of Liver Fibrosis in Patients with Chronic Hepatitis C. Exp Ther Med 2020, 20, 228, doi:10.3892/etm.2020.9358.
- Douam, F.; Lavillette, D.; Cosset, F.-L. The Mechanism of HCV Entry into Host Cells. Prog Mol Biol Transl Sci 2015, 129, 63–107, doi:10.1016/bs.pmbts.2014.10.003.
- Hemler, M.E. Tetraspanin Functions and Associated Microdomains. Nat. Rev. Mol. Cell Biol. 2005, 6, 801–811, doi:10.1038/nrm1736.
- Berditchevski, F. Complexes of Tetraspanins with Integrins: More than Meets the Eye. J. Cell. Sci. 2001, 114, 4143–4151.
- Alisi, A.; Arciello, M.; Petrini, S.; Conti, B.; Missale, G.; Balsano, C. Focal Adhesion Kinase (FAK) Mediates the Induction of pro-Oncogenic and Fibrogenic Phenotypes in Hepatitis C Virus (HCV)-Infected Cells. PLoS ONE 2012, 7, e44147, doi:10.1371/journal.pone.0044147.
- Vicente-Manzanares, M.; Webb, D.J.; Horwitz, A.R. Cell Migration at a Glance. J. Cell. Sci. 2005, 118, 4917–4919, doi:10.1242/jcs.02662.
- Martínez, S.M.; Crespo, G.; Navasa, M.; Forns, X. Noninvasive Assessment of Liver Fibrosis. Hepatology 2011, 53, 325–335, doi:10.1002/hep.24013.
- Karsdal, M.A.; Manon-Jensen, T.; Genovese, F.; Kristensen, J.H.; Nielsen, M.J.; Sand, J.M.B.; Hansen, N.-U.B.; Bay-Jensen, A.-C.; Bager, C.L.; Krag, A.; et al. Novel Insights into the Function and Dynamics of Extracellular Matrix in Liver Fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G807-830, doi:10.1152/ajpgi.00447.2014.
- Ferrell, L. Liver Pathology: Cirrhosis, Hepatitis, and Primary Liver Tumors. Update and Diagnostic Problems. Mod. Pathol. 2000, 13, 679–704, doi:10.1038/modpathol.3880119.
- Govaere, O.; Cockell, S.; Van Haele, M.; Wouters, J.; Van Delm, W.; Van den Eynde, K.; Bianchi, A.; van Eijsden, R.; Van Steenbergen, W.; Monbaliu, D.; et al. High-Throughput Sequencing Identifies Aetiology-Dependent Differences in Ductular Reaction in Human Chronic Liver Disease. J. Pathol. 2019, 248, 66–76, doi:10.1002/path.5228.
- Trivedi, S.; Murthy, S.; Sharma, H.; Hartlage, A.S.; Kumar, A.; Gadi, S.; Simmonds, P.; Chauhan, L.V.; Scheel, T.K.H.; Billerbeck, E.; et al. Viral Persistence, Liver Disease and Host Response in Hepatitis C-like Virus Rat Model. Hepatology 2017, doi:10.1002/hep.29494.
- Khatun, M.; Ray, R.B. Mechanisms Underlying Hepatitis C Virus-Associated Hepatic Fibrosis. Cells 2019, 8, doi:10.3390/cells8101249.
- Nielsen, M.J.; Karsdal, M.A.; Kazankov, K.; Grønbaek, H.; Krag, A.; Leeming, D.J.; Schuppan, D.; George, J. Fibrosis Is Not Just Fibrosis - Basement Membrane Modelling and Collagen Metabolism Differs between Hepatitis B- and C-Induced Injury. Aliment. Pharmacol. Ther. 2016, 44, 1242–1252, doi:10.1111/apt.13819.
- Guido, M.; Mangia, A.; Faa, G.; Gruppo Italiano Patologi Apparato Digerente (GIPAD); Società Italiana di Anatomia Patologica e Citopatologia Diagnostica/International Academy of Pathology, Italian division (SIAPEC/IAP) Chronic Viral Hepatitis: The Histology Report. Dig Liver Dis 2011, 43 Suppl 4, S331-343, doi:10.1016/S1590-8658(11)60589-6.
- Beltra, J.-C.; Decaluwe, H. Cytokines and Persistent Viral Infections. Cytokine 2016, 82, 4–15, doi:10.1016/j.cyto.2016.02.006.
- Govaere, O.; Petz, M.; Wouters, J.; Vandewynckel, Y.-P.; Scott, E.J.; Topal, B.; Nevens, F.; Verslype, C.; Anstee, Q.M.; Van Vlierberghe, H.; et al. The PDGFRα-Laminin B1-Keratin 19 Cascade Drives Tumor Progression at the Invasive Front of Human Hepatocellular Carcinoma. Oncogene 2017, 36, 6605–6616, doi:10.1038/onc.2017.260.
- Martinez-Quetglas, I.; Pinyol, R.; Dauch, D.; Torrecilla, S.; Tovar, V.; Moeini, A.; Alsinet, C.; Portela, A.; Rodriguez-Carunchio, L.; Solé, M.; et al. IGF2 Is Up-Regulated by Epigenetic Mechanisms in Hepatocellular Carcinomas and Is an Actionable Oncogene Product in Experimental Models. Gastroenterology 2016, 151, 1192–1205, doi:10.1053/j.gastro.2016.09.001.
- Preisser, L.; Miot, C.; Le Guillou-Guillemette, H.; Beaumont, E.; Foucher, E.D.; Garo, E.; Blanchard, S.; Frémaux, I.; Croué, A.; Fouchard, I.; et al. IL-34 and Macrophage Colony-Stimulating Factor Are Overexpressed in Hepatitis C Virus Fibrosis and Induce Profibrotic Macrophages That Promote Collagen Synthesis by Hepatic Stellate Cells. Hepatology 2014, 60, 1879–1890, doi:10.1002/hep.27328.
- Tummala, K.S.; Brandt, M.; Teijeiro, A.; Graña, O.; Schwabe, R.F.; Perna, C.; Djouder, N. Hepatocellular Carcinomas Originate Predominantly from Hepatocytes and Benign Lesions from Hepatic Progenitor Cells. Cell Rep 2017, 19, 584–600, doi:10.1016/j.celrep.2017.03.059.
- Schulze-Krebs, A.; Preimel, D.; Popov, Y.; Bartenschlager, R.; Lohmann, V.; Pinzani, M.; Schuppan, D. Hepatitis C Virus-Replicating Hepatocytes Induce Fibrogenic Activation of Hepatic Stellate Cells. Gastroenterology 2005, 129, 246–258.
- Shin, J.Y.; Hur, W.; Wang, J.S.; Jang, J.W.; Kim, C.W.; Bae, S.H.; Jang, S.K.; Yang, S.-H.; Sung, Y.C.; Kwon, O.-J.; et al. HCV Core Protein Promotes Liver Fibrogenesis via Up-Regulation of CTGF with TGF-Beta1. Exp Mol Med 2005, 37, 138–145, doi:10.1038/emm.2005.19.
- Baiocchini, A.; Montaldo, C.; Conigliaro, A.; Grimaldi, A.; Correani, V.; Mura, F.; Ciccosanti, F.; Rotiroti, N.; Brenna, A.; Montalbano, M.; et al. Extracellular Matrix Molecular Remodeling in Human Liver Fibrosis Evolution. PLoS ONE 2016, 11, e0151736, doi:10.1371/journal.pone.0151736.
- Mormone, E.; Lu, Y.; Ge, X.; Fiel, M.I.; Nieto, N. Fibromodulin, an Oxidative Stress-Sensitive Proteoglycan, Regulates the Fibrogenic Response to Liver Injury in Mice. Gastroenterology 2012, 142, 612-621.e5, doi:10.1053/j.gastro.2011.11.029.
- Benzoubir, N.; Lejamtel, C.; Battaglia, S.; Testoni, B.; Benassi, B.; Gondeau, C.; Perrin-Cocon, L.; Desterke, C.; Thiers, V.; Samuel, D.; et al. HCV Core-Mediated Activation of Latent TGF-β via Thrombospondin Drives the Crosstalk between Hepatocytes and Stromal Environment. J. Hepatol. 2013, 59, 1160–1168, doi:10.1016/j.jhep.2013.07.036.
- Bataller, R.; Paik, Y.; Lindquist, J.N.; Lemasters, J.J.; Brenner, D.A. Hepatitis C Virus Core and Nonstructural Proteins Induce Fibrogenic Effects in Hepatic Stellate Cells. Gastroenterology 2004, 126, 529–540, doi:10.1053/j.gastro.2003.11.018.
- Núñez, O.; Fernández-Martínez, A.; Majano, P.L.; Apolinario, A.; Gómez-Gonzalo, M.; Benedicto, I.; López-Cabrera, M.; Boscá, L.; Clemente, G.; García-Monzón, C.; et al. Increased Intrahepatic Cyclooxygenase 2, Matrix Metalloproteinase 2, and Matrix Metalloproteinase 9 Expression Is Associated with Progressive Liver Disease in Chronic Hepatitis C Virus Infection: Role of Viral Core and NS5A Proteins. Gut 2004, 53, 1665–1672, doi:10.1136/gut.2003.038364.
- Iqbal, J.; Sarkar-Dutta, M.; McRae, S.; Ramachandran, A.; Kumar, B.; Waris, G. Osteopontin Regulates Hepatitis C Virus (HCV) Replication and Assembly by Interacting with HCV Proteins and Lipid Droplets and by Binding to Receptors ΑVβ3 and CD44. J. Virol. 2018, 92, doi:10.1128/JVI.02116-17.
- Shirasaki, T.; Honda, M.; Yamashita, T.; Nio, K.; Shimakami, T.; Shimizu, R.; Nakasyo, S.; Murai, K.; Shirasaki, N.; Okada, H.; et al. The Osteopontin-CD44 Axis in Hepatic Cancer Stem Cells Regulates IFN Signaling and HCV Replication. Scientific Reports 2018, 8, 13143, doi:10.1038/s41598-018-31421-6.
- Jee, M.H.; Hong, K.Y.; Park, J.H.; Lee, J.S.; Kim, H.S.; Lee, S.H.; Jang, S.K. New Mechanism of Hepatic Fibrogenesis: Hepatitis C Virus Infection Induces Transforming Growth Factor Β1 Production through Glucose-Regulated Protein 94. J. Virol. 2015, 90, 3044–3055, doi:10.1128/JVI.02976-15.
- Chida, T.; Ito, M.; Nakashima, K.; Kanegae, Y.; Aoshima, T.; Takabayashi, S.; Kawata, K.; Nakagawa, Y.; Yamamoto, M.; Shimano, H.; et al. Critical Role of CREBH-Mediated Induction of Transforming Growth Factor Β2 by Hepatitis C Virus Infection in Fibrogenic Responses in Hepatic Stellate Cells. Hepatology 2017, 66, 1430–1443, doi:10.1002/hep.29319.
- Kwon, Y.-C.; Sasaki, R.; Meyer, K.; Ray, R. Hepatitis C Virus Core Protein Modulates Endoglin (CD105) Signaling Pathway for Liver Pathogenesis. J. Virol. 2017, 91, doi:10.1128/JVI.01235-17.
- Xue, Y.; Mars, W.M.; Bowen, W.; Singhi, A.D.; Stoops, J.; Michalopoulos, G.K. Hepatitis C Virus Mimics Effects of Glypican-3 on CD81 and Promotes Development of Hepatocellular Carcinomas via Activation of Hippo Pathway in Hepatocytes. Am J Pathol 2018, 188, 1469–1477, doi:10.1016/j.ajpath.2018.02.013.
- Kim, H.; Bose, S.K.; Meyer, K.; Ray, R. Hepatitis C Virus Impairs Natural Killer Cell-Mediated Augmentation of Complement Synthesis. J. Virol. 2014, 88, 2564–2571, doi:10.1128/JVI.02988-13.
- Lu, L.; Zhang, Q.; Wu, K.; Chen, X.; Zheng, Y.; Zhu, C.; Wu, J. Hepatitis C Virus NS3 Protein Enhances Cancer Cell Invasion by Activating Matrix Metalloproteinase-9 and Cyclooxygenase-2 through ERK/P38/NF-ΚB Signal Cascade. Cancer Lett. 2015, 356, 470–478, doi:10.1016/j.canlet.2014.09.027.
- Presser, L.D.; Haskett, A.; Waris, G. Hepatitis C Virus-Induced Furin and Thrombospondin-1 Activate TGF-Β1: Role of TGF-Β1 in HCV Replication. Virology 2011, 412, 284–296, doi:10.1016/j.virol.2010.12.051.
- Sakata, K.; Hara, M.; Terada, T.; Watanabe, N.; Takaya, D.; Yaguchi, S.; Matsumoto, T.; Matsuura, T.; Shirouzu, M.; Yokoyama, S.; et al. HCV NS3 Protease Enhances Liver Fibrosis via Binding to and Activating TGF-β Type I Receptor. Sci Rep 2013, 3, 3243, doi:10.1038/srep03243.
- Wen, C.; He, X.; Ma, H.; Hou, N.; Wei, C.; Song, T.; Zhang, Y.; Sun, L.; Ma, Q.; Zhong, H. Hepatitis C Virus Infection Downregulates the Ligands of the Activating Receptor NKG2D. Cell. Mol. Immunol. 2008, 5, 475–478, doi:10.1038/cmi.2008.60.
- Verga-Gérard, A.; Porcherot, M.; Meyniel-Schicklin, L.; André, P.; Lotteau, V.; Perrin-Cocon, L. Hepatitis C Virus/Human Interactome Identifies SMURF2 and the Viral Protease as Critical Elements for the Control of TGF-β Signaling. FASEB J. 2013, 27, 4027–4040, doi:10.1096/fj.13-229187.
- Li, Y.; Zhang, Q.; Liu, Y.; Luo, Z.; Kang, L.; Qu, J.; Liu, W.; Xia, X.; Liu, Y.; Wu, K.; et al. Hepatitis C Virus Activates Bcl-2 and MMP-2 Expression through Multiple Cellular Signaling Pathways. J. Virol. 2012, 86, 12531–12543, doi:10.1128/JVI.01136-12.
- Chusri, P.; Kumthip, K.; Hong, J.; Zhu, C.; Duan, X.; Jilg, N.; Fusco, D.N.; Brisac, C.; Schaefer, E.A.; Cai, D.; et al. HCV Induces Transforming Growth Factor Β1 through Activation of Endoplasmic Reticulum Stress and the Unfolded Protein Response. Sci Rep 2016, 6, 22487, doi:10.1038/srep22487.
- Choi, S.-H.; Hwang, S.B. Modulation of the Transforming Growth Factor-Beta Signal Transduction Pathway by Hepatitis C Virus Nonstructural 5A Protein. J Biol Chem 2006, 281, 7468–7478, doi:10.1074/jbc.M512438200.
- Naba, A.; Clauser, K.R.; Hoersch, S.; Liu, H.; Carr, S.A.; Hynes, R.O. The Matrisome: In Silico Definition and in Vivo Characterization by Proteomics of Normal and Tumor Extracellular Matrices. Mol. Cell Proteomics 2012, 11, M111.014647, doi:10.1074/mcp.M111.014647.
- Naba, A.; Clauser, K.R.; Whittaker, C.A.; Carr, S.A.; Tanabe, K.K.; Hynes, R.O. Extracellular Matrix Signatures of Human Primary Metastatic Colon Cancers and Their Metastases to Liver. BMC Cancer 2014, 14, 518, doi:10.1186/1471-2407-14-518.
- Decaris, M.L.; Emson, C.L.; Li, K.; Gatmaitan, M.; Luo, F.; Cattin, J.; Nakamura, C.; Holmes, W.E.; Angel, T.E.; Peters, M.G.; et al. Turnover Rates of Hepatic Collagen and Circulating Collagen-Associated Proteins in Humans with Chronic Liver Disease. PloS One 2015, 10, e0123311, doi:10.1371/journal.pone.0123311.
- Asselah, T.; Bièche, I.; Laurendeau, I.; Paradis, V.; Vidaud, D.; Degott, C.; Martinot, M.; Bedossa, P.; Valla, D.; Vidaud, M.; et al. Liver Gene Expression Signature of Mild Fibrosis in Patients with Chronic Hepatitis C. Gastroenterology 2005, 129, 2064–2075, doi:10.1053/j.gastro.2005.09.010.
- Yasui, Y.; Abe, T.; Kurosaki, M.; Matsunaga, K.; Higuchi, M.; Tamaki, N.; Watakabe, K.; Okada, M.; Wang, W.; Shimizu, T.; et al. Non-Invasive Liver Fibrosis Assessment Correlates with Collagen and Elastic Fiber Quantity in Patients with Hepatitis C Virus Infection. Hepatol Res 2019, 49, 33–41, doi:10.1111/hepr.13286.
- Bracht, T.; Schweinsberg, V.; Trippler, M.; Kohl, M.; Ahrens, M.; Padden, J.; Naboulsi, W.; Barkovits, K.; Megger, D.A.; Eisenacher, M.; et al. Analysis of Disease-Associated Protein Expression Using Quantitative Proteomics—Fibulin-5 Is Expressed in Association with Hepatic Fibrosis. J. Proteome Res. 2015, 14, 2278–2286, doi:10.1021/acs.jproteome.5b00053.
- Nielsen, M.J.; Veidal, S.S.; Karsdal, M.A.; Ørsnes-Leeming, D.J.; Vainer, B.; Gardner, S.D.; Hamatake, R.; Goodman, Z.D.; Schuppan, D.; Patel, K. Plasma Pro-C3 (N-Terminal Type III Collagen Propeptide) Predicts Fibrosis Progression in Patients with Chronic Hepatitis C. Liver Int. 2015, 35, 429–437, doi:10.1111/liv.12700.
- Murawaki, Y.; Ikuta, Y.; Koda, M.; Kawasaki, H. Serum Type III Procollagen Peptide, Type IV Collagen 7S Domain, Central Triple-Helix of Type IV Collagen and Tissue Inhibitor of Metalloproteinases in Patients with Chronic Viral Liver Disease: Relationship to Liver Histology. Hepatology 1994, 20, 780–787.
- Valva, P.; Casciato, P.; Diaz Carrasco, J.M.; Gadano, A.; Galdame, O.; Galoppo, M.C.; Mullen, E.; De Matteo, E.; Preciado, M.V. The Role of Serum Biomarkers in Predicting Fibrosis Progression in Pediatric and Adult Hepatitis C Virus Chronic Infection. PLoS One 2011, 6, e23218, doi:10.1371/journal.pone.0023218.
- Lichtinghagen, R.; Michels, D.; Haberkorn, C.I.; Arndt, B.; Bahr, M.; Flemming, P.; Manns, M.P.; Boeker, K.H. Matrix Metalloproteinase (MMP)-2, MMP-7, and Tissue Inhibitor of Metalloproteinase-1 Are Closely Related to the Fibroproliferative Process in the Liver during Chronic Hepatitis C. J. Hepatol. 2001, 34, 239–247, doi:10.1016/s0168-8278(00)00037-4.
- Ljumovic, D.; Diamantis, I.; Alegakis, A.K.; Kouroumalis, E.A. Differential Expression of Matrix Metalloproteinases in Viral and Non-Viral Chronic Liver Diseases. Clin. Chim. Acta 2004, 349, 203–211, doi:10.1016/j.cccn.2004.06.028.
- Martinez-Castillo, M.; Hernandez-Barragan, A.; Flores-Vasconcelos, I.; Galicia-Moreno, M.; Rosique-Oramas, D.; Perez-Hernandez, J.L.; Higuera-De la Tijera, F.; Montalvo-Jave, E.E.; Torre-Delgadillo, A.; Cordero-Perez, P.; et al. Production and Activity of Matrix Metalloproteinases during Liver Fibrosis Progression of Chronic Hepatitis C Patients. World J Hepatol 2021, 13, 218–232, doi:10.4254/wjh.v13.i2.218.
- Murawaki, Y.; Ikuta, Y.; Kawasaki, H. Clinical Usefulness of Serum Tissue Inhibitor of Metalloproteinases (TIMP)-2 Assay in Patients with Chronic Liver Disease in Comparison with Serum TIMP-1. Clin. Chim. Acta 1999, 281, 109–120.
- Dudás, J.; Kovalszky, I.; Gallai, M.; Nagy, J.O.; Schaff, Z.; Knittel, T.; Mehde, M.; Neubauer, K.; Szalay, F.; Ramadori, G. Expression of Decorin, Transforming Growth Factor-Beta 1, Tissue Inhibitor Metalloproteinase 1 and 2, and Type IV Collagenases in Chronic Hepatitis. American Journal of Clinical Pathology 2001, 115, 725–735, doi:10.1309/J8CD-E9C8-X4NG-GTVG.
- Ramnath, D.; Irvine, K.M.; Lukowski, S.W.; Horsfall, L.U.; Loh, Z.; Clouston, A.D.; Patel, P.J.; Fagan, K.J.; Iyer, A.; Lampe, G.; et al. Hepatic Expression Profiling Identifies Steatosis-Independent and Steatosis-Driven Advanced Fibrosis Genes. JCI insight 2018, 3, doi:10.1172/jci.insight.120274.
- Dong, C.; Li, H.-J.; Chang, S.; Liao, H.-J.; Zhang, Z.-P.; Huang, P.; Tang, H.-H. A Disintegrin and Metalloprotease with Thrombospondin Motif 2 May Contribute to Cirrhosis in Humans through the Transforming Growth Factor-β/SMAD Pathway. Gut Liver 2013, 7, 213–220, doi:10.5009/gnl.2013.7.2.213.
- Kuhn, J.; Gressner, O.A.; Götting, C.; Gressner, A.M.; Kleesiek, K. Increased Serum Xylosyltransferase Activity in Patients with Liver Fibrosis. Clin. Chim. Acta 2009, 409, 123–126, doi:10.1016/j.cca.2009.09.013.
- Gressner, O.A.; Gao, C. Monitoring Fibrogenic Progression in the Liver. Clin. Chim. Acta 2014, 433, 111–122, doi:10.1016/j.cca.2014.02.021.
- Zhu, Z.W.; Friess, H.; Wang, L.; Abou-Shady, M.; Zimmermann, A.; Lander, A.D.; Korc, M.; Kleeff, J.; Büchler, M.W. Enhanced Glypican-3 Expression Differentiates the Majority of Hepatocellular Carcinomas from Benign Hepatic Disorders. Gut 2001, 48, 558–564, doi:10.1136/gut.48.4.558.
- Llovet, J.M.; Chen, Y.; Wurmbach, E.; Roayaie, S.; Fiel, M.I.; Schwartz, M.; Thung, S.N.; Khitrov, G.; Zhang, W.; Villanueva, A.; et al. A Molecular Signature to Discriminate Dysplastic Nodules from Early Hepatocellular Carcinoma in HCV Cirrhosis. Gastroenterology 2006, 131, 1758–1767, doi:10.1053/j.gastro.2006.09.014.
- Wurmbach, E.; Chen, Y.; Khitrov, G.; Zhang, W.; Roayaie, S.; Schwartz, M.; Fiel, I.; Thung, S.; Mazzaferro, V.; Bruix, J.; et al. Genome-Wide Molecular Profiles of HCV-Induced Dysplasia and Hepatocellular Carcinoma. Hepatology 2007, 45, 938–947, doi:10.1002/hep.21622.
- Clemente, M.; Núñez, O.; Lorente, R.; Rincón, D.; Matilla, A.; Salcedo, M.; Catalina, M.V.; Ripoll, C.; Iacono, O.L.; Bañares, R.; et al. Increased Intrahepatic and Circulating Levels of Endoglin, a TGF-Beta1 Co-Receptor, in Patients with Chronic Hepatitis C Virus Infection: Relationship to Histological and Serum Markers of Hepatic Fibrosis. J Viral Hepat 2006, 13, 625–632, doi:10.1111/j.1365-2893.2006.00733.x.
- Guéchot, J.; Laudat, A.; Loria, A.; Serfaty, L.; Poupon, R.; Giboudeau, J. Diagnostic Accuracy of Hyaluronan and Type III Procollagen Amino-Terminal Peptide Serum Assays as Markers of Liver Fibrosis in Chronic Viral Hepatitis C Evaluated by ROC Curve Analysis. Clin Chem 1996, 42, 558–563.
- Karsdal, M.A.; Daniels, S.J.; Holm Nielsen, S.; Bager, C.; Rasmussen, D.G.K.; Loomba, R.; Surabattula, R.; Villesen, I.F.; Luo, Y.; Shevell, D.; et al. Collagen Biology and Non-Invasive Biomarkers of Liver Fibrosis. Liver Int. 2020, 40, 736–750, doi:10.1111/liv.14390.
- Taleb, R.S.Z.; Moez, P.; Younan, D.; Eisenacher, M.; Tenbusch, M.; Sitek, B.; Bracht, T. Quantitative Proteome Analysis of Plasma Microparticles for the Characterization of HCV-Induced Hepatic Cirrhosis and Hepatocellular Carcinoma. Proteomics Clin Appl 2017, 11, doi:10.1002/prca.201700014.
- El-Karef, A.; Kaito, M.; Tanaka, H.; Ikeda, K.; Nishioka, T.; Fujita, N.; Inada, H.; Adachi, Y.; Kawada, N.; Nakajima, Y.; et al. Expression of Large Tenascin-C Splice Variants by Hepatic Stellate Cells/Myofibroblasts in Chronic Hepatitis C. J Hepatol 2007, 46, 664–673, doi:10.1016/j.jhep.2006.10.011.
- Benbow, J.H.; Elam, A.D.; Bossi, K.L.; Massengill, D.L.; Brandon-Warner, E.; Anderson, W.E.; Culberson, C.R.; Russo, M.W.; deLemos, A.S.; Schrum, L.W. Analysis of Plasma Tenascin-C in Post-HCV Cirrhosis: A Prospective Study. Dig Dis Sci 2018, 63, 653–664, doi:10.1007/s10620-017-4860-z.
- Choi, S.S.; Claridge, L.C.; Jhaveri, R.; Swiderska-Syn, M.; Clark, P.; Suzuki, A.; Pereira, T.A.; Mi, Z.; Kuo, P.C.; Guy, C.D.; et al. Osteopontin Is Up-Regulated in Chronic Hepatitis C and Is Associated with Cellular Permissiveness for Hepatitis C Virus Replication. Clin Sci (Lond) 2014, 126, 845–855, doi:10.1042/CS20130473.
- Urtasun, R.; Lopategi, A.; George, J.; Leung, T.-M.; Lu, Y.; Wang, X.; Ge, X.; Fiel, M.I.; Nieto, N. Osteopontin, an Oxidant Stress Sensitive Cytokine, up-Regulates Collagen-I via Integrin α(V)β(3) Engagement and PI3K/PAkt/NFκB Signaling. Hepatology 2012, 55, 594–608, doi:10.1002/hep.24701.
- Hackl, N.J.; Bersch, C.; Feick, P.; Antoni, C.; Franke, A.; Singer, M.V.; Nakchbandi, I.A. Circulating Fibronectin Isoforms Predict the Degree of Fibrosis in Chronic Hepatitis C. Scand J Gastroenterol 2010, 45, 349–356, doi:10.3109/00365520903490606.
- Bracht, T.; Mölleken, C.; Ahrens, M.; Poschmann, G.; Schlosser, A.; Eisenacher, M.; Stühler, K.; Meyer, H.E.; Schmiegel, W.H.; Holmskov, U.; et al. Evaluation of the Biomarker Candidate MFAP4 for Non-Invasive Assessment of Hepatic Fibrosis in Hepatitis C Patients. J Transl Med 2016, 14, 1–9, doi:10.1186/s12967-016-0952-3.
- Mölleken, C.; Ahrens, M.; Schlosser, A.; Dietz, J.; Eisenacher, M.; Meyer, H.E.; Schmiegel, W.; Holmskov, U.; Sarrazin, C.; Sorensen, G.L.; et al. Direct-Acting Antivirals-Based Therapy Decreases Hepatic Fibrosis Serum Biomarker Microfibrillar-Associated Protein 4 in Hepatitis C Patients. Clin Mol Hepatol 2018, 25, 42–51, doi:10.3350/cmh.2018.0029.
- Castilla, A.; Prieto, J.; Fausto, N. Transforming Growth Factors Beta 1 and Alpha in Chronic Liver Disease. Effects of Interferon Alfa Therapy. N Engl J Med 1991, 324, 933–940, doi:10.1056/NEJM199104043241401.
- Kinnman, N.; Andersson, U.; Hultcrantz, R. In Situ Expression of Transforming Growth Factor-Beta1-3, Latent Transforming Growth Factor-Beta Binding Protein and Tumor Necrosis Factor-Alpha in Liver Tissue from Patients with Chronic Hepatitis C. Scand J Gastroenterol 2000, 35, 1294–1300, doi:10.1080/003655200453656.
- Divella, R.; Daniele, A.; Gadaleta, C.; Tufaro, A.; Venneri, M.T.; Paradiso, A.; Quaranta, M. Circulating Transforming Growth Factor-β and Epidermal Growth Factor Receptor as Related to Virus Infection in Liver Carcinogenesis. Anticancer Res 2012, 32, 141–145.
- Bader, H.L.; Lambert, E.; Guiraud, A.; Malbouyres, M.; Driever, W.; Koch, M.; Ruggiero, F. Zebrafish Collagen XIV Is Transiently Expressed in Epithelia and Is Required for Proper Function of Certain Basement Membranes. J. Biol. Chem. 2013, 288, 6777–6787, doi:10.1074/jbc.M112.430637.
- Grässel, S.; Unsöld, C.; Schäcke, H.; Bruckner-Tuderman, L.; Bruckner, P. Collagen XVI Is Expressed by Human Dermal Fibroblasts and Keratinocytes and Is Associated with the Microfibrillar Apparatus in the Upper Papillary Dermis. Matrix Biol. 1999, 18, 309–317, doi:10.1016/s0945-053x(99)00019-0.
- Uitto, J.; Pulkkinen, L. Molecular Complexity of the Cutaneous Basement Membrane Zone. Mol. Biol. Rep. 1996, 23, 35–46, doi:10.1007/BF00357071.
- Schuppan, D.; Cramer, T.; Bauer, M.; Strefeld, T.; Hahn, E.G.; Herbst, H. Hepatocytes as a Source of Collagen Type XVIII Endostatin. Lancet 1998, 352, 879–880, doi:10.1016/S0140-6736(05)60006-2.
- Heljasvaara, R.; Aikio, M.; Ruotsalainen, H.; Pihlajaniemi, T. Collagen XVIII in Tissue Homeostasis and Dysregulation - Lessons Learned from Model Organisms and Human Patients. Matrix Biol. 2017, 57–58, 55–75, doi:10.1016/j.matbio.2016.10.002.
- Vadasz, Z.; Kessler, O.; Akiri, G.; Gengrinovitch, S.; Kagan, H.M.; Baruch, Y.; Izhak, O.B.; Neufeld, G. Abnormal Deposition of Collagen around Hepatocytes in Wilson’s Disease Is Associated with Hepatocyte Specific Expression of Lysyl Oxidase and Lysyl Oxidase like Protein-2. J. Hepatol. 2005, 43, 499–507, doi:10.1016/j.jhep.2005.02.052.
- Barry-Hamilton, V.; Spangler, R.; Marshall, D.; McCauley, S.; Rodriguez, H.M.; Oyasu, M.; Mikels, A.; Vaysberg, M.; Ghermazien, H.; Wai, C.; et al. Allosteric Inhibition of Lysyl Oxidase-like-2 Impedes the Development of a Pathologic Microenvironment. Nat. Med. 2010, 16, 1009–1017, doi:10.1038/nm.2208.
- Liu, S.B.; Ikenaga, N.; Peng, Z.-W.; Sverdlov, D.Y.; Greenstein, A.; Smith, V.; Schuppan, D.; Popov, Y. Lysyl Oxidase Activity Contributes to Collagen Stabilization during Liver Fibrosis Progression and Limits Spontaneous Fibrosis Reversal in Mice. FASEB J. 2016, 30, 1599–1609, doi:10.1096/fj.14-268425.
- Ikenaga, N.; Peng, Z.-W.; Vaid, K.A.; Liu, S.B.; Yoshida, S.; Sverdlov, D.Y.; Mikels-Vigdal, A.; Smith, V.; Schuppan, D.; Popov, Y.V. Selective Targeting of Lysyl Oxidase-like 2 (LOXL2) Suppresses Hepatic Fibrosis Progression and Accelerates Its Reversal. Gut 2017, 66, 1697–1708, doi:10.1136/gutjnl-2016-312473.
- Giovannini, C.; Fornari, F.; Indio, V.; Trerè, D.; Renzulli, M.; Vasuri, F.; Cescon, M.; Ravaioli, M.; Perrucci, A.; Astolfi, A.; et al. Direct Antiviral Treatments for Hepatitis C Virus Have Off-Target Effects of Oncologic Relevance in Hepatocellular Carcinoma. Cancers (Basel) 2020, 12, doi:10.3390/cancers12092674.
- Fontana, R.J.; Dienstag, J.L.; Bonkovsky, H.L.; Sterling, R.K.; Naishadham, D.; Goodman, Z.D.; Lok, A.S.F.; Wright, E.C.; Su, G.L.; HALT-C Trial Group Serum Fibrosis Markers Are Associated with Liver Disease Progression in Non-Responder Patients with Chronic Hepatitis C. Gut 2010, 59, 1401–1409, doi:10.1136/gut.2010.207423.
- Medeiros, T.; Saraiva, G.N.; Moraes, L.A.; Gomes, A.C.; Lacerda, G.S.; Leite, P.E.; Esberard, E.B.; Andrade, T.G.; Xavier, A.R.; Quírico-Santos, T.; et al. Liver Fibrosis Improvement in Chronic Hepatitis C after Direct Acting-Antivirals Is Accompanied by Reduced Profibrogenic Biomarkers-a Role for MMP-9/TIMP-1. Dig Liver Dis 2020, doi:10.1016/j.dld.2020.05.004.
- Schwettmann, L.; Wehmeier, M.; Jokovic, D.; Aleksandrova, K.; Brand, K.; Manns, M.P.; Lichtinghagen, R.; Bahr, M.J. Hepatic Expression of A Disintegrin and Metalloproteinase (ADAM) and ADAMs with Thrombospondin Motives (ADAM-TS) Enzymes in Patients with Chronic Liver Diseases. J. Hepatol. 2008, 49, 243–250, doi:10.1016/j.jhep.2008.03.020.
- Bourd-Boittin, K.; Basset, L.; Bonnier, D.; L’helgoualc’h, A.; Samson, M.; Théret, N. CX3CL1/Fractalkine Shedding by Human Hepatic Stellate Cells: Contribution to Chronic Inflammation in the Liver. J. Cell. Mol. Med. 2009, 13, 1526–1535, doi:10.1111/j.1582-4934.2009.00787.x.
- Parkes, J.; Guha, I.N.; Roderick, P.; Harris, S.; Cross, R.; Manos, M.M.; Irving, W.; Zaitoun, A.; Wheatley, M.; Ryder, S.; et al. Enhanced Liver Fibrosis (ELF) Test Accurately Identifies Liver Fibrosis in Patients with Chronic Hepatitis C. J. Viral Hepat. 2011, 18, 23–31, doi:10.1111/j.1365-2893.2009.01263.x.
- Kumar, V.; Kato, N.; Urabe, Y.; Takahashi, A.; Muroyama, R.; Hosono, N.; Otsuka, M.; Tateishi, R.; Omata, M.; Nakagawa, H.; et al. Genome-Wide Association Study Identifies a Susceptibility Locus for HCV-Induced Hepatocellular Carcinoma. Nat. Genet. 2011, 43, 455–458, doi:10.1038/ng.809.
- Zwirner, N.W.; Dole, K.; Stastny, P. Differential Surface Expression of MICA by Endothelial Cells, Fibroblasts, Keratinocytes, and Monocytes. Hum. Immunol. 1999, 60, 323–330, doi:10.1016/s0198-8859(98)00128-1.
- Goto, K.; Kato, N. MICA SNPs and the NKG2D System in Virus-Induced HCC. J. Gastroenterol. 2015, 50, 261–272, doi:10.1007/s00535-014-1000-9.
- Waldhauer, I.; Goehlsdorf, D.; Gieseke, F.; Weinschenk, T.; Wittenbrink, M.; Ludwig, A.; Stevanovic, S.; Rammensee, H.-G.; Steinle, A. Tumor-Associated MICA Is Shed by ADAM Proteases. Cancer Res. 2008, 68, 6368–6376, doi:10.1158/0008-5472.CAN-07-6768.
- Kohga, K.; Takehara, T.; Tatsumi, T.; Ishida, H.; Miyagi, T.; Hosui, A.; Hayashi, N. Sorafenib Inhibits the Shedding of Major Histocompatibility Complex Class I-Related Chain A on Hepatocellular Carcinoma Cells by down-Regulating a Disintegrin and Metalloproteinase 9. Hepatology 2010, 51, 1264–1273, doi:10.1002/hep.23456.
- Kohga, K.; Takehara, T.; Tatsumi, T.; Miyagi, T.; Ishida, H.; Ohkawa, K.; Kanto, T.; Hiramatsu, N.; Hayashi, N. Anticancer Chemotherapy Inhibits MHC Class I-Related Chain a Ectodomain Shedding by Downregulating ADAM10 Expression in Hepatocellular Carcinoma. Cancer Res. 2009, 69, 8050–8057, doi:10.1158/0008-5472.CAN-09-0789.
- Goto, K.; Arai, J.; Stephanou, A.; Kato, N. Novel Therapeutic Features of Disulfiram against Hepatocellular Carcinoma Cells with Inhibitory Effects on a Disintegrin and Metalloproteinase 10. Oncotarget 2018, 9, 18821–18831, doi:10.18632/oncotarget.24568.
- Huang, C.-F.; Huang, C.-Y.; Yeh, M.-L.; Wang, S.-C.; Chen, K.-Y.; Ko, Y.-M.; Lin, C.-C.; Tsai, Y.-S.; Tsai, P.-C.; Lin, Z.-Y.; et al. Genetics Variants and Serum Levels of MHC Class I Chain-Related A in Predicting Hepatocellular Carcinoma Development in Chronic Hepatitis C Patients Post Antiviral Treatment. EBioMedicine 2017, 15, 81–89, doi:10.1016/j.ebiom.2016.11.031.
- Theocharis, A.D.; Skandalis, S.S.; Tzanakakis, G.N.; Karamanos, N.K. Proteoglycans in Health and Disease: Novel Roles for Proteoglycans in Malignancy and Their Pharmacological Targeting. FEBS J. 2010, 277, 3904–3923, doi:10.1111/j.1742-4658.2010.07800.x.
- Shi, Q.; Jiang, J.; Luo, G. Syndecan-1 Serves as the Major Receptor for Attachment of Hepatitis C Virus to the Surfaces of Hepatocytes. J. Virol. 2013, 87, 6866–6875, doi:10.1128/JVI.03475-12.
- Lefèvre, M.; Felmlee, D.J.; Parnot, M.; Baumert, T.F.; Schuster, C. Syndecan 4 Is Involved in Mediating HCV Entry through Interaction with Lipoviral Particle-Associated Apolipoprotein E. PLoS ONE 2014, 9, e95550, doi:10.1371/journal.pone.0095550.
- Matsumoto, A.; Ono, M.; Fujimoto, Y.; Gallo, R.L.; Bernfield, M.; Kohgo, Y. Reduced Expression of Syndecan-1 in Human Hepatocellular Carcinoma with High Metastatic Potential. Int. J. Cancer 1997, 74, 482–491.
- Regős, E.; Karászi, K.; Reszegi, A.; Kiss, A.; Schaff, Z.; Baghy, K.; Kovalszky, I. Syndecan-1 in Liver Diseases. Pathol. Oncol. Res. 2020, 26, 813–819, doi:10.1007/s12253-019-00617-0.
- Pönighaus, C.; Ambrosius, M.; Casanova, J.C.; Prante, C.; Kuhn, J.; Esko, J.D.; Kleesiek, K.; Götting, C. Human Xylosyltransferase II Is Involved in the Biosynthesis of the Uniform Tetrasaccharide Linkage Region in Chondroitin Sulfate and Heparan Sulfate Proteoglycans. J. Biol. Chem. 2007, 282, 5201–5206, doi:10.1074/jbc.M611665200.
- Baghy, K.; Tátrai, P.; Regős, E.; Kovalszky, I. Proteoglycans in Liver Cancer. World Journal of Gastroenterology 2016, 22, 379–393, doi:10.3748/wjg.v22.i1.379.
- Liu, B.; Paranjpe, S.; Bowen, W.C.; Bell, A.W.; Luo, J.-H.; Yu, Y.-P.; Mars, W.M.; Michalopoulos, G.K. Investigation of the Role of Glypican 3 in Liver Regeneration and Hepatocyte Proliferation. Am. J. Pathol. 2009, 175, 717–724, doi:10.2353/ajpath.2009.081129.
- Toretsky, J.A.; Zitomersky, N.L.; Eskenazi, A.E.; Voigt, R.W.; Strauch, E.D.; Sun, C.C.; Huber, R.; Meltzer, S.J.; Schlessinger, D. Glypican-3 Expression in Wilms Tumor and Hepatoblastoma. Journal of Pediatric Hematology/Oncology 2001, 23, 496–499, doi:10.1097/00043426-200111000-00006.
- Bhave, V.S.; Mars, W.; Donthamsetty, S.; Zhang, X.; Tan, L.; Luo, J.; Bowen, W.C.; Michalopoulos, G.K. Regulation of Liver Growth by Glypican 3, CD81, Hedgehog, and Hhex. Am. J. Pathol. 2013, 183, 153–159, doi:10.1016/j.ajpath.2013.03.013.
- Zvibel, I.; Halfon, P.; Fishman, S.; Penaranda, G.; Leshno, M.; Or, A.B.; Halpern, Z.; Oren, R. Syndecan 1 (CD138) Serum Levels: A Novel Biomarker in Predicting Liver Fibrosis Stage in Patients with Hepatitis C. Liver International: Official Journal of the International Association for the Study of the Liver 2009, 29, 208–212, doi:10.1111/j.1478-3231.2008.01830.x.
- Shimizu, Y.; Mizuno, S.; Fujinami, N.; Suzuki, T.; Saito, K.; Konishi, M.; Takahashi, S.; Gotohda, N.; Tada, T.; Toyoda, H.; et al. Plasma and Tumoral Glypican-3 Levels Are Correlated in Patients with Hepatitis C Virus-Related Hepatocellular Carcinoma. Cancer Sci. 2020, 111, 334–342, doi:10.1111/cas.14251.
- Tsuchiya, N.; Sawada, Y.; Endo, I.; Saito, K.; Uemura, Y.; Nakatsura, T. Biomarkers for the Early Diagnosis of Hepatocellular Carcinoma. World Journal of Gastroenterology 2015, 21, 10573–10583, doi:10.3748/wjg.v21.i37.10573.
- Sun, C.K.; Chua, M.-S.; He, J.; So, S.K. Suppression of Glypican 3 Inhibits Growth of Hepatocellular Carcinoma Cells through Up-Regulation of TGF-Β2. Neoplasia 2011, 13, 735–747.
- Regős, E.; Abdelfattah, H.H.; Reszegi, A.; Szilák, L.; Werling, K.; Szabó, G.; Kiss, A.; Schaff, Z.; Kovalszky, I.; Baghy, K. Syndecan-1 Inhibits Early Stages of Liver Fibrogenesis by Interfering with TGFβ1 Action and Upregulating MMP14. Matrix Biol. 2018, 68–69, 474–489, doi:10.1016/j.matbio.2018.02.008.
- Skandalis, S.S.; Karalis, T.T.; Chatzopoulos, A.; Karamanos, N.K. Hyaluronan-CD44 Axis Orchestrates Cancer Stem Cell Functions. Cellular Signalling 2019, 63, 109377, doi:10.1016/j.cellsig.2019.109377.
- Park, N.R.; Cha, J.H.; Jang, J.W.; Bae, S.H.; Jang, B.; Kim, J.-H.; Hur, W.; Choi, J.Y.; Yoon, S.K. Synergistic Effects of CD44 and TGF-Β1 through AKT/GSK-3β/β-Catenin Signaling during Epithelial-Mesenchymal Transition in Liver Cancer Cells. Biochemical and Biophysical Research Communications 2016, 477, 568–574, doi:10.1016/j.bbrc.2016.06.077.
- Castelli, G.; Pelosi, E.; Testa, U. Liver Cancer: Molecular Characterization, Clonal Evolution and Cancer Stem Cells. Cancers 2017, 9, doi:10.3390/cancers9090127.
- Kakehashi, A.; Ishii, N.; Sugihara, E.; Gi, M.; Saya, H.; Wanibuchi, H. CD44 Variant 9 Is a Potential Biomarker of Tumor Initiating Cells Predicting Survival Outcome in Hepatitis C Virus-Positive Patients with Resected Hepatocellular Carcinoma. Cancer Science 2016, 107, 609–618, doi:10.1111/cas.12908.
- Abe, T.; Fukuhara, T.; Wen, X.; Ninomiya, A.; Moriishi, K.; Maehara, Y.; Takeuchi, O.; Kawai, T.; Akira, S.; Matsuura, Y. CD44 Participates in IP-10 Induction in Cells in Which Hepatitis C Virus RNA Is Replicating, through an Interaction with Toll-like Receptor 2 and Hyaluronan. Journal of Virology 2012, 86, 6159–6170, doi:10.1128/JVI.06872-11.
- Lord, M.S.; Tang, F.; Rnjak-Kovacina, J.; Smith, J.G.W.; Melrose, J.; Whitelock, J.M. The Multifaceted Roles of Perlecan in Fibrosis. Matrix Biol 2018, 68–69, 150–166, doi:10.1016/j.matbio.2018.02.013.
- Batmunkh, E.; Tátrai, P.; Szabó, E.; Lódi, C.; Holczbauer, A.; Páska, C.; Kupcsulik, P.; Kiss, A.; Schaff, Z.; Kovalszky, I. Comparison of the Expression of Agrin, a Basement Membrane Heparan Sulfate Proteoglycan, in Cholangiocarcinoma and Hepatocellular Carcinoma. Hum Pathol 2007, 38, 1508–1515, doi:10.1016/j.humpath.2007.02.017.
- Binder, M.J.; McCoombe, S.; Williams, E.D.; McCulloch, D.R.; Ward, A.C. The Extracellular Matrix in Cancer Progression: Role of Hyalectan Proteoglycans and ADAMTS Enzymes. Cancer Letters 2017, 385, 55–64, doi:10.1016/j.canlet.2016.11.001.
- Sobhy, A.; Fakhry M, M.; A Azeem, H.; Ashmawy, A.M.; Omar Khalifa, H. Significance of Biglycan and Osteopontin as Non-Invasive Markers of Liver Fibrosis in Patients with Chronic Hepatitis B Virus and Chronic Hepatitis C Virus. J Investig Med 2019, 67, 681–685, doi:10.1136/jim-2018-000840.
- Roedig, H.; Damiescu, R.; Zeng-Brouwers, J.; Kutija, I.; Trebicka, J.; Wygrecka, M.; Schaefer, L. Danger Matrix Molecules Orchestrate CD14/CD44 Signaling in Cancer Development. Seminars in Cancer Biology 2020, 62, 31–47, doi:10.1016/j.semcancer.2019.07.026.
- Baghy, K.; Iozzo, R.V.; Kovalszky, I. Decorin-TGFβ Axis in Hepatic Fibrosis and Cirrhosis. The Journal of Histochemistry and Cytochemistry: Official Journal of the Histochemistry Society 2012, 60, 262–268, doi:10.1369/0022155412438104.
- Ivanov, A.V.; Bartosch, B.; Smirnova, O.A.; Isaguliants, M.G.; Kochetkov, S.N. HCV and Oxidative Stress in the Liver. Viruses 2013, 5, 439–469, doi:10.3390/v5020439.
- Campello, E.; Radu, C.M.; Zanetto, A.; Bulato, C.; Shalaby, S.; Spiezia, L.; Franceschet, E.; Burra, P.; Russo, F.P.; Simioni, P. Changes in Plasma Circulating Microvesicles in Patients with HCV-Related Cirrhosis after Treatment with Direct-Acting Antivirals. Liver International: Official Journal of the International Association for the Study of the Liver 2020, 40, 913–920, doi:10.1111/liv.14234.
- Gerarduzzi, C.; Hartmann, U.; Leask, A.; Drobetsky, E. The Matrix Revolution: Matricellular Proteins and Restructuring of the Cancer Microenvironment. Cancer Res 2020, 80, 2705–2717, doi:10.1158/0008-5472.CAN-18-2098.
- Ramazani, Y.; Knops, N.; Elmonem, M.A.; Nguyen, T.Q.; Arcolino, F.O.; van den Heuvel, L.; Levtchenko, E.; Kuypers, D.; Goldschmeding, R. Connective Tissue Growth Factor (CTGF) from Basics to Clinics. Matrix Biol 2018, 68–69, 44–66, doi:10.1016/j.matbio.2018.03.007.
- Tanaka, H.; El-Karef, A.; Kaito, M.; Kinoshita, N.; Fujita, N.; Horiike, S.; Watanabe, S.; Yoshida, T.; Adachi, Y. Circulating Level of Large Splice Variants of Tenascin-C Is a Marker of Piecemeal Necrosis Activity in Patients with Chronic Hepatitis C. Liver Int 2006, 26, 311–318, doi:10.1111/j.1478-3231.2005.01229.x.
- Coombes, J.D.; Swiderska-Syn, M.; Dollé, L.; Reid, D.; Eksteen, B.; Claridge, L.; Briones-Orta, M.A.; Shetty, S.; Oo, Y.H.; Riva, A.; et al. Osteopontin Neutralisation Abrogates the Liver Progenitor Cell Response and Fibrogenesis in Mice. Gut 2015, 64, 1120–1131, doi:10.1136/gutjnl-2013-306484.
- Bruha, R.; Vitek, L.; Smid, V. Osteopontin - A Potential Biomarker of Advanced Liver Disease. Ann Hepatol 2020, 19, 344–352, doi:10.1016/j.aohep.2020.01.001.
- Matsue, Y.; Tsutsumi, M.; Hayashi, N.; Saito, T.; Tsuchishima, M.; Toshikuni, N.; Arisawa, T.; George, J. Serum Osteopontin Predicts Degree of Hepatic Fibrosis and Serves as a Biomarker in Patients with Hepatitis C Virus Infection. PLoS One 2015, 10, e0118744, doi:10.1371/journal.pone.0118744.
- Athwal, V.S.; Pritchett, J.; Martin, K.; Llewellyn, J.; Scott, J.; Harvey, E.; Zaitoun, A.M.; Mullan, A.F.; Zeef, L.A.H.; Friedman, S.L.; et al. Publisher Correction: SOX9 Regulated Matrix Proteins Are Increased in Patients Serum and Correlate with Severity of Liver Fibrosis. Sci Rep 2019, 9, 11547, doi:10.1038/s41598-019-47715-2.
- Wang, X.; Lopategi, A.; Ge, X.; Lu, Y.; Kitamura, N.; Urtasun, R.; Leung, T.-M.; Fiel, M.I.; Nieto, N. Osteopontin Induces Ductular Reaction Contributing to Liver Fibrosis. Gut 2014, 63, 1805–1818, doi:10.1136/gutjnl-2013-306373.
- Sia, D.; Villanueva, A.; Friedman, S.L.; Llovet, J.M. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology 2017, 152, 745–761, doi:10.1053/j.gastro.2016.11.048.
- Iso, Y.; Sawada, T.; Okada, T.; Kubota, K. Loss of E-Cadherin MRNA and Gain of Osteopontin MRNA Are Useful Markers for Detecting Early Recurrence of HCV-Related Hepatocellular Carcinoma. J Surg Oncol 2005, 92, 304–311, doi:10.1002/jso.20388.
- Mochida, S.; Hashimoto, M.; Matsui, A.; Naito, M.; Inao, M.; Nagoshi, S.; Nagano, M.; Egashira, T.; Mishiro, S.; Fujiwara, K. Genetic Polymorphims in Promoter Region of Osteopontin Gene May Be a Marker Reflecting Hepatitis Activity in Chronic Hepatitis C Patients. Biochem Biophys Res Commun 2004, 313, 1079–1085, doi:10.1016/j.bbrc.2003.12.045.
- Gressner, O.A.; Gressner, A.M. Connective Tissue Growth Factor: A Fibrogenic Master Switch in Fibrotic Liver Diseases. Liver Int 2008, 28, 1065–1079, doi:10.1111/j.1478-3231.2008.01826.x.
- Paradis, V.; Dargere, D.; Vidaud, M.; De Gouville, A.C.; Huet, S.; Martinez, V.; Gauthier, J.M.; Ba, N.; Sobesky, R.; Ratziu, V.; et al. Expression of Connective Tissue Growth Factor in Experimental Rat and Human Liver Fibrosis. Hepatology 1999, 30, 968–976, doi:10.1002/hep.510300425.
- Hora, C.; Negro, F.; Leandro, G.; Oneta, C.M.; Rubbia-Brandt, L.; Muellhaupt, B.; Helbling, B.; Malinverni, R.; Gonvers, J.-J.; Dufour, J.-F.; et al. Connective Tissue Growth Factor, Steatosis and Fibrosis in Patients with Chronic Hepatitis C. Liver Int 2008, 28, 370–376, doi:10.1111/j.1478-3231.2007.01608.x.
- Nagaraja, T.; Chen, L.; Balasubramanian, A.; Groopman, J.E.; Ghoshal, K.; Jacob, S.T.; Leask, A.; Brigstock, D.R.; Anand, A.R.; Ganju, R.K. Activation of the Connective Tissue Growth Factor (CTGF)-Transforming Growth Factor β 1 (TGF-β 1) Axis in Hepatitis C Virus-Expressing Hepatocytes. PLoS One 2012, 7, e46526, doi:10.1371/journal.pone.0046526.
- Williams, M.J.; Clouston, A.D.; Forbes, S.J. Links between Hepatic Fibrosis, Ductular Reaction, and Progenitor Cell Expansion. Gastroenterology 2014, 146, 349–356, doi:10.1053/j.gastro.2013.11.034.
- Vasel, M.; Rutz, R.; Bersch, C.; Feick, P.; Singer, M.V.; Kirschfink, M.; Nakchbandi, I.A. Complement Activation Correlates with Liver Necrosis and Fibrosis in Chronic Hepatitis C. Clin Immunol 2014, 150, 149–156, doi:10.1016/j.clim.2013.11.014.
- Lorenzini, S.; Bird, T.G.; Boulter, L.; Bellamy, C.; Samuel, K.; Aucott, R.; Clayton, E.; Andreone, P.; Bernardi, M.; Golding, M.; et al. Characterisation of a Stereotypical Cellular and Extracellular Adult Liver Progenitor Cell Niche in Rodents and Diseased Human Liver. Gut 2010, 59, 645–654, doi:10.1136/gut.2009.182345.
- Matsumoto, S.; Yamamoto, K.; Nagano, T.; Okamoto, R.; Ibuki, N.; Tagashira, M.; Tsuji, T. Immunohistochemical Study on Phenotypical Changes of Hepatocytes in Liver Disease with Reference to Extracellular Matrix Composition. Liver 1999, 19, 32–38, doi:10.1111/j.1478-3231.1999.tb00006.x.
- Kanta, J.; Dooley, S.; Delvoux, B.; Breuer, S.; D’Amico, T.; Gressner, A.M. Tropoelastin Expression Is Up-Regulated during Activation of Hepatic Stellate Cells and in the Livers of CCl(4)-Cirrhotic Rats. Liver 2002, 22, 220–227.
- Zhu, S.; Ye, L.; Bennett, S.; Xu, H.; He, D.; Xu, J. Molecular Structure and Function of Microfibrillar-Associated Proteins in Skeletal and Metabolic Disorders and Cancers. J Cell Physiol 2020, doi:10.1002/jcp.29893.
- Papke, C.L.; Yanagisawa, H. Fibulin-4 and Fibulin-5 in Elastogenesis and beyond: Insights from Mouse and Human Studies. Matrix Biol 2014, 37, 142–149, doi:10.1016/j.matbio.2014.02.004.
- Kanzaki, T.; Olofsson, A.; Morén, A.; Wernstedt, C.; Hellman, U.; Miyazono, K.; Claesson-Welsh, L.; Heldin, C.H. TGF-Beta 1 Binding Protein: A Component of the Large Latent Complex of TGF-Beta 1 with Multiple Repeat Sequences. Cell 1990, 61, 1051–1061, doi:10.1016/0092-8674(90)90069-q.
- Kusakabe, M.; Cheong, P.-L.; Nikfar, R.; McLennan, I.S.; Koishi, K. The Structure of the TGF-Beta Latency Associated Peptide Region Determines the Ability of the Proprotein Convertase Furin to Cleave TGF-Betas. J Cell Biochem 2008, 103, 311–320, doi:10.1002/jcb.21407.
- Fausto, N.; Mead, J.E.; Gruppuso, P.A.; Castilla, A.; Jakowlew, S.B. Effects of TGF-Beta s in the Liver: Cell Proliferation and Fibrogenesis. Ciba Found. Symp. 1991, 157, 165–174; discussion 174-177.
- Li, G.; Jiang, Q.; Xu, K. CREB Family: A Significant Role in Liver Fibrosis. Biochimie 2019, 163, 94–100, doi:10.1016/j.biochi.2019.05.014.
- Lua, I.; Li, Y.; Zagory, J.A.; Wang, K.S.; French, S.W.; Sévigny, J.; Asahina, K. Characterization of Hepatic Stellate Cells, Portal Fibroblasts, and Mesothelial Cells in Normal and Fibrotic Livers. J. Hepatol. 2016, 64, 1137–1146, doi:10.1016/j.jhep.2016.01.010.
- Kataria, Y.; Deaton, R.J.; Enk, E.; Jin, M.; Petrauskaite, M.; Dong, L.; Goldenberg, J.R.; Cotler, S.J.; Jensen, D.M.; van Breemen, R.B.; et al. Retinoid and Carotenoid Status in Serum and Liver among Patients at High-Risk for Liver Cancer. BMC Gastroenterol 2016, 16, 30, doi:10.1186/s12876-016-0432-5.
- Fabregat, I.; Moreno-Càceres, J.; Sánchez, A.; Dooley, S.; Dewidar, B.; Giannelli, G.; Ten Dijke, P.; IT-LIVER Consortium TGF-β Signalling and Liver Disease. FEBS J 2016, 283, 2219–2232, doi:10.1111/febs.13665.
- Sasaki, R.; Devhare, P.; Ray, R.B.; Ray, R. Hepatitis C Virus-Induced Tumor-Initiating Cancer Stem-like Cells Activate Stromal Fibroblasts in a Xenograft Tumor Model. Hepatology 2017, 66, 1766–1778, doi:10.1002/hep.29346.
- Nelson, D.R.; Gonzalez-Peralta, R.P.; Qian, K.; Xu, Y.; Marousis, C.G.; Davis, G.L.; Lau, J.Y. Transforming Growth Factor-Beta 1 in Chronic Hepatitis C. J Viral Hepat 1997, 4, 29–35, doi:10.1046/j.1365-2893.1997.00124.x.
- Taniguchi, H.; Kato, N.; Otsuka, M.; Goto, T.; Yoshida, H.; Shiratori, Y.; Omata, M. Hepatitis C Virus Core Protein Upregulates Transforming Growth Factor-Beta 1 Transcription. J Med Virol 2004, 72, 52–59, doi:10.1002/jmv.10545.
- Miyanari, Y.; Atsuzawa, K.; Usuda, N.; Watashi, K.; Hishiki, T.; Zayas, M.; Bartenschlager, R.; Wakita, T.; Hijikata, M.; Shimotohno, K. The Lipid Droplet Is an Important Organelle for Hepatitis C Virus Production. Nat. Cell Biol. 2007, 9, 1089–1097, doi:10.1038/ncb1631.
- Waris, G.; Tardif, K.D.; Siddiqui, A. Endoplasmic Reticulum (ER) Stress: Hepatitis C Virus Induces an ER-Nucleus Signal Transduction Pathway and Activates NF-KappaB and STAT-3. Biochem Pharmacol 2002, 64, 1425–1430, doi:10.1016/s0006-2952(02)01300-x.
- Meurer, S.K.; Alsamman, M.; Scholten, D.; Weiskirchen, R. Endoglin in Liver Fibrogenesis: Bridging Basic Science and Clinical Practice. World J Biol Chem 2014, 5, 180–203, doi:10.4331/wjbc.v5.i2.180.
- About, F.; Bibert, S.; Jouanguy, E.; Nalpas, B.; Lorenzo, L.; Rattina, V.; Zarhrate, M.; Hanein, S.; Munteanu, M.; Müllhaupt, B.; et al. Identification of an Endoglin Variant Associated With HCV-Related Liver Fibrosis Progression by Next-Generation Sequencing. Front Genet 2019, 10, 1024, doi:10.3389/fgene.2019.01024.
- Xu, L.; Hui, A.Y.; Albanis, E.; Arthur, M.J.; O’Byrne, S.M.; Blaner, W.S.; Mukherjee, P.; Friedman, S.L.; Eng, F.J. Human Hepatic Stellate Cell Lines, LX-1 and LX-2: New Tools for Analysis of Hepatic Fibrosis. Gut 2005, 54, 142–151, doi:10.1136/gut.2004.042127.
- Florimond, A.; Chouteau, P.; Bruscella, P.; Le Seyec, J.; Mérour, E.; Ahnou, N.; Mallat, A.; Lotersztajn, S.; Pawlotsky, J.-M. Human Hepatic Stellate Cells Are Not Permissive for Hepatitis C Virus Entry and Replication. Gut 2015, 64, 957–965, doi:10.1136/gutjnl-2013-305634.
- Aoudjehane, L.; Bisch, G.; Scatton, O.; Granier, C.; Gaston, J.; Housset, C.; Roingeard, P.; Cosset, F.-L.; Perdigao, F.; Balladur, P.; et al. Infection of Human Liver Myofibroblasts by Hepatitis C Virus: A Direct Mechanism of Liver Fibrosis in Hepatitis C. PLoS ONE 2015, 10, e0134141, doi:10.1371/journal.pone.0134141.
- Devhare, P.B.; Sasaki, R.; Shrivastava, S.; Di Bisceglie, A.M.; Ray, R.; Ray, R.B. Exosome-Mediated Intercellular Communication between Hepatitis C Virus-Infected Hepatocytes and Hepatic Stellate Cells. J Virol 2017, 91, doi:10.1128/JVI.02225-16.
- Bartosch, B. Piecing Together the Key Players of Fibrosis in Chronic Hepatitis C: What Roles Do Non-Hepatic Liver Resident Cell Types Play? Gut 2015, 64, 862–863, doi:10.1136/gutjnl-2014-307957.
- Foschi, F.G.; Domenicali, M.; Giacomoni, P.; Dall’Aglio, A.C.; Conti, F.; Borghi, A.; Bevilacqua, V.; Napoli, L.; Mirici, F.; Cucchetti, A.; et al. Is There an Association between Commonly Employed Biomarkers of Liver Fibrosis and Liver Stiffness in the General Population? Ann Hepatol 2020, 19, 380–387, doi:10.1016/j.aohep.2020.04.003.
- Meissner, E.G.; McLaughlin, M.; Matthews, L.; Gharib, A.M.; Wood, B.J.; Levy, E.; Sinkus, R.; Virtaneva, K.; Sturdevant, D.; Martens, C.; et al. Simtuzumab Treatment of Advanced Liver Fibrosis in HIV and HCV-Infected Adults: Results of a 6-Month Open-Label Safety Trial. Liver Int. 2016, 36, 1783–1792, doi:10.1111/liv.13177.
- Roehlen, N.; Crouchet, E.; Baumert, T.F. Liver Fibrosis: Mechanistic Concepts and Therapeutic Perspectives. Cells 2020, 9, doi:10.3390/cells9040875.
This entry is adapted from the peer-reviewed paper 10.3390/cancers13092270
