Perilipins are a family of five closely related proteins expressed on the surface of lipid droplets (LD) in several tissues acting in several pathways involved in lipid metabolism. Recent studies have shown that Plin5 depletion acts protectively in the pathogenesis of liver injury underpinning the importance of pathways associated with PLIN5. PLIN5 expression is involved in pro-inflammatory cytokine regulation and mitochondrial damage, as well as endoplasmic reticulum (ER) stress, making it critical target of the NAFLD-HCC studies.
- perilipin 5
- non-alcoholic liver disease
- fatty liver
- hepatocellular carcinoma
- cancer
1. Introduction
Lipid accumulation in the liver plays a pivotal role in the pathogenesis of NAFLD that might result from dysfunction of cellular lipid trafficking and lipid droplet formation. The accepted model of LD formation is based on the biosynthesis of neutral lipids (e.g., triacylglycerides, TAGs) by esterification of fatty acids (FA) into diacylglycerol or sterols [1][2] Figure 1 [1].
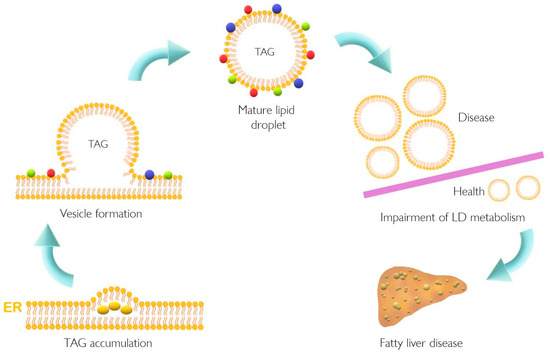
Figure 1. Lipid droplet formation and excessive accumulation leads to hepatic pathological conditions. TAG accumulation in the ER membrane, accompanied by the action of different proteins, leads to LD formation. This excessive accumulation results in impairment in storage of LD in several tissues resulting in pathological conditions such as non-alcoholic fatty liver disease. Colorful spheres around LD represent perilipins, Rab proteins, and Arf-COPI proteins. ER, endoplasmic reticulum; LD, lipid droplet; TAG, triacylglyceride.
NAFLD is characterized by augmented accumulation of hepatic LD, which is in line with increased PLIN5 expression [3]. Recently, alterations in lipid metabolism have also been proposed as a new hallmark of cancer [4][5][6]. Moreover, several cellular processes such as oxidative stress, ER stress, and inflammation have been related to non-alcoholic steatohepatitis (NASH) the progressed form of NAFLD [7][8] as well as to cancer development and progression [9][10]. Meanwhile, PLIN5 has indeed been implicated in the regulation of lipid and glucose homeostasis and many cellular processes such as oxidative stress, ER stress, inflammation, and autophagy [3][11][12][13][14][15][16][17][18][19][20][21]. This evidence once again supports the notion that PLIN5 could play a direct role in transition from NAFLD to HCC.
2. Plin5 Transcriptional Regulation and Interactions
In one study, PLIN5 was named myocardial LD protein (MLDP) due to its cardial expression [22]. The other two groups reported the expression of PLIN5 in oxidative tissues-enriched PAT protein (OXPAT) and lipid storage droplet protein 5 (LSDP5), a protein not exclusively expressed in the heart, but highly expressed in oxidative tissues, such as muscle and liver [23][24]. Peroxisome proliferator-activated receptor (PPAR) was shown to regulate the transcription of Plin5 since the first intron of the gene contains a PPAR regulatory element (PPRE) [23]. All three groups showed that Plin5 is induced in the liver under fasting conditions in a PPARα-dependent fashion [23][22][24].
Different enzymes, such as adipose triglyceride lipase (ATGL), hormone-sensitive lipase (HSL), and monoglyceride lipase (MGL) regulate lipid hydrolysis. TAG hydrolysis is catalyzed by ATGL, followed by cleavage of one molecule of FA forming diaglycerides by HSL and finally hydrolysis is completed by MGL [1]. The interaction between PLIN5 and ATGL and its protein activator, α-β-hydrolase domain-containing 5 (ABHD5), results in decreased lipolysis [25][26]. In this scenario, different lipid droplet proteins play a key role in regulating lipid metabolism and TAG storage in cells.
3. The Role of PLIN5 in Lipid Metabolism and NAFLD
One of the first studies to elucidate the role of PLIN5 in hepatocytes demonstrated that PLIN5 expression is induced by FFA in a time-independent manner [27]. Additionally, a study performed in Plin5-deficient mouse hepatocyte AML-12 cells showed a decrease in cellular TAG content as well as an increase of mitochondrial oxidation [28]. To gain a better understanding of consequences resulting from Plin5 depletion in vivo, several knockout (KO) models in mice have been developed. Table 1 contains key points of the major models reported as well as the feeding conditions evaluated in each of these, underpinning the notion that the role of PLIN5 in metabolism seems to be tissue-specific.
Table 1. Phenotypes of Plin5 transgenic mouse models.
| Model | Tissue | Findings | References |
|---|---|---|---|
| Plin5−/− mice | Heart | No LD formation in fed and fasting state ↓ TAG content in fed and fasting state |
[29] |
| Soleus muscle | ↓ TAG content | ||
| Liver | ↓ TAG content in fed state ↑ TAG content in fasting state |
||
| BAT | ↓ TAG content in fed and fasting state | ||
| WAT | ↓ TAG content in fasting mice | ||
| Plin5−/− mice | Whole body | ↑ Carbohydrate oxidation | [11] |
| Muscle | ↑ Skeletal muscle insulin resistance | ||
| Liver | Improvement of insulin sensitivity | ||
| Plin5−/− mice | Heart | ↓ Cardiac LD formation ↑ Cardiac FA oxidation |
[30] |
| Plin5−/− mice | Liver | ↓ Hepatic TAG content ↑ Lipolysis ↑ Mitochondrial proliferation ↑ Mitochondrial oxidative capacity ↑ Expression of pro-inflammatory genes under an HFD ↑ Expression of ER stress-related genes ↑ Lipid peroxidation |
[14] |
| Plin5−/− mice | Liver | Under HFD vs. CtrD: ↓ Fat scoring ↓ Ballooning of hepatocytes ↓ Levels of liver damage enzymes ALP, ALT, AST ↓ Bilirubin levels ↓ Cholesterol levels ↑ Levels of mitochondrial structure and trafficking markers ↑ Lipogenesis ↓ Inflammatory markers ↓ Levels of arachidonic acid |
[18] |
| MKO mice | Skeletal muscle | ↑ Fat mass ↓ Respiratory exchange ratio ↑ FA oxidation under HFD ↑ Oxidative stress ↑ TAG content ↓ Pro-inflammatory markers |
[16] |
| Heart | ↓ TAG content No ER stress, inflammation and oxidative stress |
||
| Hepatocyte-specific Plin5−/− mice |
Liver | ↓ FA consumption ↓ FA oxidation ↓ TAG secretion ↓ Lipid peroxidation and oxidative stress Insulin resistance and enhancement under an HFD Glucose intolerance under HFD TAG accumulation under HFD |
[12] |
| CM-Plin5 mice | Heart | ↑ Accumulation of TAGs ↑ TAG hydrolytic activities: ↑ ATGL and CGI-58 protein levels Moderately reduced FA oxidizing gene expression levels |
[31] |
| MCK-Plin5 mice | Skeletal muscle | ↑ LD formation ↓ Body weight compared to non-transgenic littermates under control and HFD diet ↑ Expression of ER stress markers |
[32] |
| Heart | ↑ TAG content | ||
| Diaphragm | ↑ TAG content | ||
| EDL | ↑ TAG content | ||
| Gastrocnemius | ↑ TAG content | ||
| Liver | ↓ Cholesterol levels in an HFD compared to control diet littermates ↓ Lipid uptake ↓ Inflammatory markers |
ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BAT, brown adipose tissue; CtrD, control diet; CM-Plin5, cardiac muscle-specific overexpression of Plin5; EDL, extensor digitorum longus (muscle); ER, endoplasmic reticulum; FA, fatty acid; HFD, high fat diet; LD, lipid droplet; MCK-Plin5, skeletal muscle-specific overexpression of Plin5; MKO, muscle-specific Plin5 knockout; Plin5−/−, Plin5 deficient; TAG, triacylglyceride; WAT, white adipose tissue.
4. PLIN5 Is Responsive to Cellular Processes Altered in NAFLD
NAFLD is a condition characterized of dyslipidemia, increased oxidative stress, ROS production, and mitochondrial alterations provoking inflammatory responses [8]. There are several reports analyzing how PLIN5 participates in each of these processes. In one study, the depletion of Plin5 in an in vivo mouse model under fasting conditions was associated with decreased LD formation and TAG content in the heart [29]. The same effect was observed in soleus muscle, BAT, and WAT.
PLIN5 showed implications in the inflammatory response in different models, and strikingly, LCN2, a well-known acute pro-inflammatory protein and critical regulator of lipid uptake, has been shown to act as a regulator of PLIN5 activity in a NASH model based in a methionine- and choline-deficient diet [33]. This report supports that LCN2 impacts PLIN5 expression in a PPAR-independent manner, since its stimulation with an PPAR-γ agonist failed to suppress its expression [33].
Furthermore, studies on Plin5−/− mice have revealed a close local connection of mitochondria and LDs [18][34]. In the respective model, the oxidative capacity of mitochondria in cardiac muscle was impaired due to changes in lipid composition of the membrane and its polarization [34]. Super-resolution microscopy-based approaches in skeletal muscle have shown that under basal conditions there are three pools of PLIN5, namely in the cytosol, in close proximity of mitochondria, and surrounding LDs in the mitochondria–LD interface [35]. Moreover, it was observed that increased FA levels led to an augment of PLIN5 in the mitochondria–LD interface pool, suggesting that this relocation dynamic of PLIN5 might facilitate flux of FA from LDs to mitochondria [35][36]. All these data demonstrate the fine-tuned regulation of PLIN5 in lipotoxicity, inflammation response, and mitochondrial dynamics, processes that are all closely related in the development of NAFLD. The major biological processes in which PLIN5 participates are illustrated in Figure 3.
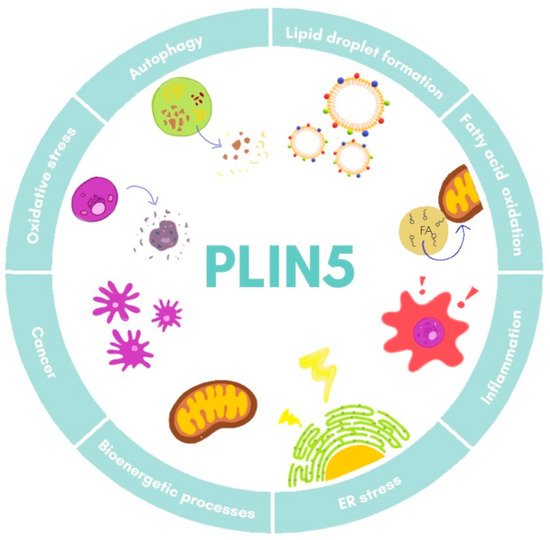
Figure 3. Cellular functions of PLIN5. Major processes in which PLIN5 has been reported to participate are shown. ER, endoplasmic reticulum.
5. Perilipins in Cancer: The Unrevealed Role of PLIN5
Information contained in the Human Protein Atlas indicates PLIN5 level alterations in some types of cancer (http://www.proteinatlas.org/, last accessed 24 March 2021). A first study about participation of perilipins in neoplastic steatogenesis was reported by Straub and colleagues. They suggested that PLIN1, PLIN2, and PLIN3 may be involved in cancers such as hepatocellular adenoma and carcinoma, sebaceous adenoma and carcinoma, and lipoma tumors, while PLIN5 was excluded as not sufficient proof was given [37].
Little is understood so far about how PLIN5 is involved in the development or progress of cancer, but a novel transcriptome analysis of lung cancer, predicted that higher expression of the guanine nucleotide-binding protein α-inhibiting activity polypeptide 3 (GNAI3) and the lncRNA AC087521.1 was associated with a short survival time, while the NHL repeat-containing protein 2 (NHLRC2) and PLIN5 correlated with longer survival times [38].
Information about PLIN5 implication in HCC is still limited. It was first shown in 2019 that PLIN5 expression is upregulated in HCC tumoral areas in murine HCC models and human HCC biopsy specimens as assessed by Western blot analysis and quantitative real time PCR [39]. RNA-seq and splicing factor (SF) data of 390 HCC cases deposited in the Cancer Genome Atlas (TCGA) data portal (https://tcgadata.nci.nih.gov/tcga/, last accessed 24 March 2021) indicate that PLIN5-46808-AT was found more common in primary sites of HCC with metastasis than primary HCC [40]. It is evident that more studies elucidating HCC development are necessary to clarify the actual function of PLIN5 in HCC.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22105284
References
- Minehira, K.; Gual, P. Role of Lipid Droplet Proteins in the Development of NAFLD and Hepatic Insulin Resistance. Non-Alcoholic Fat Liver Disease; IntechOpen: London, UK, 2018; pp. 55–77.
- Olzmann, J.A.; Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 137–155.
- Ma, S.Y.; Sun, K.S.; Zhang, M.; Zhou, X.; Zheng, X.H.; Tian, S.Y.; Liu, Y.S.; Chen, L.; Gao, X.; Ye, J.; et al. Disruption of Plin5 degradation by CMA causes lipid homeostasis imbalance in NAFLD. Liver Int. 2020, 40, 2427–2438.
- Hayes, C.N.; Zhang, P.; Chayama, K. The role of lipids in hepatocellular carcinoma. In Hepatocellular Carcinoma; Tirnitz-Parker, J.E.E., Ed.; Codon Publications: Brisbane, Australia, 2019; Chapter 5.
- Corn, K.C.; Windham, M.A.; Rafat, M. Lipids in the tumor microenvironment: From cancer progression to treatment. Prog. Lipid Res. 2020, 80.
- Calvisi, D.F.; Wang, C.; Ho, C.; Ladu, S.; Lee, S.A.; Mattu, S.; Destefanis, G.; Delogu, S.; Zimmermann, A.; Ericsson, J.; et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology 2011, 140, 1071–1083.
- Ashraf, N.U.; Sheikh, T.A. Endoplasmic reticulum stress and Oxidative stress in the pathogenesis of Non-alcoholic fatty liver disease. Free Radic. Res. 2015, 49, 1405–1418.
- Chen, Z.; Tian, R.; She, Z.; Cai, J.; Li, H. Role of oxidative stress in the pathogenesis of nonalcoholic fatty liver disease. Free Radic. Biol. Med. 2020, 20, 116–141.
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616.
- Saha, S.K.; Lee, S.B.; Won, J.; Choi, H.Y.; Kim, K.; Yang, G.M.; Dayem, A.A.; Cho, S.G. Correlation between oxidative stress, nutrition, and cancer initiation. Int. J. Mol. Sci. 2017, 18, 1544.
- Mason, R.R.; Mokhtar, R.; Matzaris, M.; Selathurai, A.; Kowalski, G.M.; Mokbel, N.; Meikle, P.J.; Bruce, C.R.; Watt, M.J. PLIN5 deletion remodels intracellular lipid composition and causes insulin resistance in muscle. Mol. Metab. 2014, 3, 652–663.
- Keenan, S.N.; Meex, R.C.; Lo, J.C.Y.; Ryan, A.; Nie, S.; Montgomery, M.K.; Watt, M.J. Perilipin 5 Deletion in hepatocytes remodels lipid metabolism and causes hepatic insulin resistance in mice. Diabetes 2019, 68, 543–555.
- Wang, H.; Sreenivasan, U.; Gong, D.W.; O’Connell, K.A.; Dabkowski, E.R.; Hecker, P.A.; Ionica, N.; Konig, M.; Mahurkar, A.; Sun, Y.; et al. Cardiomyocyte-specific perilipin 5 overexpression leads to myocardial steatosis and modest cardiac dysfunction. J. Lipid Res. 2013, 54, 953–965.
- Wang, C.; Zhao, Y.; Gao, X.; Li, L.; Yuan, Y.; Liu, F.; Zhang, L.; Wu, J.; Hu, P.; Zhang, X.; et al. Perilipin 5 improves hepatic lipotoxicity by inhibiting lipolysis. Hepatology 2015, 61, 870–882.
- Lin, J.; Chen, A. Perilipin 5 restores the formation of lipid droplets in activated hepatic stellate cells and inhibits their activation. Lab. Investig. 2016, 96, 791–806.
- Montgomery, M.K.; Mokhtar, R.; Bayliss, J.; Parkington, H.C.; Suturin, V.M.; Bruce, C.R.; Watt, M.J. Perilipin 5 deletion unmasks an endoplasmic reticulum Stress-Fibroblast growth factor 21 axis in skeletal muscle. Diabetes 2018, 67, 594–606.
- Tan, Y.; Jin, Y.; Wang, Q.; Huang, J.; Wu, X.; Ren, Z. Perilipin 5 protects against cellular oxidative stress by enhancing mitochondrial function in HepG2 cells. Cells 2019, 8, 1241.
- Asimakopoulou, A.; Engel, K.M.; Gassler, N.; Bracht, T.; Sitek, B.; Buhl, E.M.; Kalampoka, S.; Pinoé-Schmidt, M.; van Helden, J.; Schiller, J.; et al. Deletion of Perilipin 5 protects against hepatic injury in nonalcoholic fatty liver disease via missing inflammasome activation. Cells 2020, 9, 1346.
- Zhang, E.; Cui, W.; Lopresti, M.; Mashek, M.T.; Najt, C.P.; Hu, H.; Mashek, D.G. Hepatic PLIN5 signals via SIRT1 to promote autophagy and prevent inflammation during fasting. J. Lipid Res. 2020, 61, 338–350.
- Zhu, Y.; Ren, C.; Zhang, M.; Zhong, Y. Perilipin 5 reduces oxidative damage associated with lipotoxicity by activating the PI3K/ERK-mediated Nrf2-ARE signaling pathway in INS-1 pancreatic β-cells. Front. Endocrinol. 2020, 11, 1–14.
- Feng, J.; Xie, L.; Yu, X.; Liu, C.; Dong, H.; Lu, W.; Kong, R. Perilipin 5 ameliorates high-glucose-induced podocyte injury via Akt/GSK-3β/Nrf2-mediated suppression of apoptosis, oxidative stress, and inflammation. Biochem. Biophys. Res. Commun. 2021, 12, 22–30.
- Yamaguchi, T.; Matsushita, S.; Motojima, K.; Hirose, F.; Osumi, T. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor α. J. Biol. Chem. 2006, 281, 14232–14240.
- Dalen, K.T.; Dahl, T.; Holter, E.; Arntsen, B.; Londos, C.; Sztalryd, C. LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2007, 1771, 210–227.
- Wolins, N.E.; Quaynor, B.K.; Skinner, J.R.; Tzekov, A.; Croce, M.A.; Gropler, M.C. OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes 2006, 55, 3418–3428.
- Granneman, J.G.; Moore, H.P.H.; Mottillo, E.P.; Zhu, Z.; Zhou, L. Interactions of Perilipin-5 (Plin5) with adipose triglyceride lipase. J. Biol. Chem. 2011, 286, 5126–5135.
- Wang, H.; Bell, M.; Sreenevasan, U.; Hu, H.; Liu, J.; Dalen, K.; Londos, C.; Yamaguchi, T.; Rizzo, M.A.; Coleman, R.; et al. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J. Biol. Chem. 2011, 286, 15707–15715.
- Grasselli, E.; Voci, A.; Pesce, C.; Canesi, L.; Fugassa, E.; Gallo, G.; Vergani, L. PAT protein mRNA expression in primary rat hepatocytes: Effects of exposure to fatty acids. Int. J. Mol. Med. 2010, 25, 505–512.
- Li, H.; Song, Y.; Zhang, L.J.; Gu, Y.; Li, F.F.; Pan, S.Y. LSDP5 enhances triglyceride storage in hepatocytes by influencing lipolysis and fatty acid β-oxidation of lipid droplets. PLoS ONE 2012, 7, e36712.
- Kuramoto, K.; Okamura, T.; Yamaguchi, T.; Nakamura, T.Y.; Wakabayashi, S.; Morinaga, H.; Nomura, M.; Yanase, T.; Otsu, K.; Usuda, N.; et al. Perilipin 5, a lipid droplet-binding protein, protects heart from oxidative burden by sequestering fatty acid from excessive oxidation. J. Biol. Chem. 2012, 287, 23852–23863.
- Kimmel, A.R.; Sztalryd, C. Perilipin 5, a lipid droplet protein adapted to mitochondrial energy utilization. Curr. Opin. Lipidol. 2014, 25, 110–117.
- Pollak, N.M.; Schweiger, M.; Jaeger, D.; Kolb, D.; Kumari, M.; Schreiber, R.; Kolleritsch, S.; Markolin, P.; Grabner, G.F.; Heier, C.; et al. Cardiac-Specific overexpression of perilipin 5 provokes severe cardiac steatosis via the formation of a lipolytic barrier. J. Lipid Res. 2013, 54, 1092–1102.
- Harris, L.A.; Skinner, J.R.; Shew, T.M.; Pietka, T.A.; Abumrad, N.A.; Wolins, N.E. Perilipin 5-driven lipid droplet accumulation in skeletal muscle stimulates the expression of fibroblast growth factor 21. Diabetes 2015, 64, 2757–2768.
- Asimakopoulou, A.; Borkham-Kamphorst, E.; Henning, M.; Yagmur, E.; Gassler, N.; Liedtke, C.; Berger, T.; Mak, T.W.; Weiskirchen, R. Lipocalin-2 (LCN2) regulates PLIN5 expression and intracellular lipid droplet formation in the liver. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2014, 1841, 1513–1524.
- Andersson, L.; Drevinge, C.; Mardani, I.; Dalen, K.T.; Ståhlman, M.; Klevstig, M.; Lundqvist, A.; Haugen, F.; Adiels, M.; Fogelstrand, P.; et al. Deficiency in perilipin 5 reduces mitochondrial function and membrane depolarization in mouse hearts. Int. J. Biochem. Cell Biol. 2017, 91, 9–13.
- Gemmink, A.; Daemen, S.; Kuijpers, H.J.H.; Schaart, G.; Duimel, H.; López-Iglesias, C.; van Zandvoort, M.A.M.J.; Knoops, K.; Hesselink, M.K.C. Super-Resolution microscopy localizes perilipin 5 at lipid droplet-mitochondria interaction sites and at lipid droplets juxtaposing to perilipin 2. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 1423–1432.
- Varghese, M.; Kimler, V.A.; Ghazi, F.R.; Rathore, G.K.; Perkins, G.A.; Ellisman, M.H.; Granneman, J.G. Adipocyte lipolysis affects Perilipin 5 and cristae organization at the cardiac lipid droplet-mitochondrial interface. Sci. Rep. 2019, 9, 1–12.
- Straub, B.K.; Herpel, E.; Singer, S.; Zimbelmann, R.; Breuhahn, K.; MacHer-Goeppinger, S.; Warth, A.; Lehmann-Koch, J.; Longerich, T.; Heid, H.; et al. Lipid droplet-associated PAT-proteins show frequent and differential expression in neoplastic steatogenesis. Mod. Pathol. 2010, 23, 480–492.
- Ye, J.; Liu, H.; Xu, Z.L.; Zheng, L.; Liu, R.Y. Identification of a multidimensional transcriptome prognostic signature for lung adenocarcinoma. J. Clin. Lab. Anal. 2019, 33.
- Asimakopoulou, A.; Vucur, M.; Luedde, T.; Schneiders, S.; Kalampoka, S.; Weiss, T.S.; Weiskirchen, R. Perilipin 5 and Lipocalin 2 expression in hepatocellular carcinoma. Cancers 2019, 11, 385.
- Huang, R.; Yan, G.; Sun, H.; Zhang, J.; Song, D.; Kong, R.; Yan, P.; Hu, P.; Xie, A.; Wang, S.; et al. Identification of prognostic and metastasis-related alternative splicing signatures in hepatocellular carcinoma. Biosci. Rep. 2020, 40, 1–14.
