Nanozymes have the potential to replace natural enzymes, so they are widely used in energy conversion technologies such as biosensors and signal transduction (converting biological signals of a target into optical, electrical, or metabolic signals). The participation of nucleic acids leads nanozymes to produce richer interface effects and gives energy conversion events more attractive characteristics, creating what are called “functional nanozymes”.
- nanozyme
- nucleic acid
- modulation
- recognition
- energy conversion
1. From “Enzyme” to “Nucleic Acid Modulation for Functional Nanozyme”
1.1. Enzyme
In the field of biology, enzymes are a class of macromolecular substances with biocatalytic functions. Only under suitable temperature and acid-base conditions can enzymes change the physiological state of organisms by regulating their metabolic pathways, such as by signal transduction, gene expression, gene silencing, etc., or by obtaining new biological traits or removing certain biological traits[1][2]. Enzymes usually exhibit excellent catalytic efficiency and regio- and stereoselectivity, and they are widely used in biochemical energy conversion, playing an important role in food risk-factor detection, medical disease diagnosis and treatment, and environmental pollutant analysis[3][4].
Most natural enzymes are proteins or RNA, and some studies have pointed out that enzymes can also be DNA. Natural enzymes lose their activity when encountering nonphysiological conditions, and the preparation process of enzymes is complicated and expensive[5][6]. Therefore, in the past few decades, scientists have been seeking to synthesize compounds with properties similar to enzymes’ activity by chemical or physical methods. These compounds are stable, economical, and able to adapt to nonphysiological conditions to solve problems in practical applications[7][8].
1.2. Nanozyme
In recent years, nanoparticles (NPs) with strong tolerance to the external environment have shown dual characteristics, including biological properties and material chemical properties, and these NPs are called “nanozymes”, which are expected to replace natural enzymes[9][10]. In 2004, self-assembly triazacyclonane-functionalized thiols on the surface of gold NPs exhibited RNase-like behavior that catalyzed the cleavage of phosphate esters[11]. Then, to define this layer of gold nanoclusters, scientists proposed the concept of a “nanozyme”. NPs with enzymatic activity were later described as nanozymes[12][13]. In 2007, Yan et al. reported for the first time that Fe3O4 nanoparticle (NP) catalysts showed properties similar to peroxidase[14]. The publication of this work changed the traditional concept of inorganic nanomaterials being biologically inert substances, revealed the inherent biological effects and new characteristics of nanomaterials, and expanded from organic composites to inorganic nanomaterials, which broke the boundary between “inorganic” and “organic” in the traditional sense.
Moreover, by combining excellent physical and chemical properties with enzyme-like catalytic activity, nanozymes can realize multifunctional biological applications from detection to monitoring and treatment and have been widely studied in the fields of medicine, chemistry, food, agriculture, and environment[15]. Compared with natural enzymes, nanozymes have the following characteristics[15]:
(1) High stability: Inorganic nanomaterials are more adaptable to pH and temperature changes than natural enzymes. Some nanozyme can be used under a wide range of pH (3–12) and temperature (4–90 °C) conditions. In contrast, biological enzymes are usually easily denatured and inactivated under extreme pH and temperature conditions.
(2) Low cost: The production process of enzymes is usually complicated and expensive, while inorganic nanomaterials are easy to produce on a large scale with good catalytic activity and low cost.
(3) Recycling: Inorganic nanomaterials are recyclable, and there is no significant loss of catalytic activity in the subsequent cycles.
(4) Easy to be multifunctional: nanozymes have a large specific surface area and high surface energy and can be combined with multiple ligands to achieve multifunctionality[16][17][18][19].
Nanozymes, as new stars in science, not only have the characteristics of materials chemistry, including a large specific surface area, rich surface morphology, easy modification, and unique size and shape, but also have more attractive biological characteristics, including the ability to respond to physiological reactions and to catalyze biochemical reactions [20][21][22][23][24][25][26].
1.3. Functional Nanozyme
Due to the excellent dual properties of nanozymes, some specific studies have shown that they can respond well to the changes of biological macromolecules (proteins, nucleic acids, polysaccharides, lipids).
(1) As for proteins, Zhang et al.[27] encapsulated transition metal catalysts (TMCs) on the single-layer surface of gold nanoparticles to prepare a bio-orthogonal nanozyme. By weakening the formation of a constant protein crown (hard corona), the long-term retention of nanozyme activity in the cell is achieved.
(2) As for nucleic acids, Wang et al.[28] found that ssDNA adsorbed on g-C3N4 NSs could improve the catalytic activity of the nanosheets.
(3) As for polysaccharides, Li et al.[29] synthesized soluble molecularly imprinted nanozyme that can accurately hydrolyze the oligosaccharide maltohexaose.
(4) As for lipids, Zhang et al.[30] reported magnetic nanoparticles (iron oxide nanozyme). After a nanozyme enters the cell, it exerts peroxidase activity in the acidic environment of the lysosome, increases the level of ROS activity, destroys proteins, nucleic acids, lipids, and other biological molecules, makes them lose their functions, and kills Escherichia coli.
Overall, the structure and surface physicochemical properties of a nanozyme determines whether it has the characteristics to cope well with biological macromolecules’ changes and thus to produce unique nanobiological effects.
Among these biological macromolecules, nucleic acids have been known for specific self-assembly properties and unique molecular recognition mechanisms[31]. First, the double-stranded structure of nucleic acids has complementarity, and a series of DNA-based nanomaterials can be developed based on this complementarity between strands[32]. Moreover, as a biological recognition molecule, a nucleic acid aptamer is essentially a single-stranded DNA or RNA folded to form a specific secondary and tertiary conformation that then binds to the target molecule with high affinity and specificity[31].
The increasing development of nucleic acid technology has promoted the progress of studies on biochemical energy conversion related to biomacromolecule-modulating nanozymes (energy conversion: the energy generated by the biochemical event is transformed into other forms of energy, such as light energy, electric energy, new biological energy, or new chemical energy)[33][34][35][36][37]. Specifically, nucleobases can provide lone pairs of nitrogen and oxygen electrons, and nucleobases are an important structure of the nucleic acid phosphate backbone. Thus, nucleic acid acts as a multidentate organic ligand, which interfaces with metal ions, metal oxides, metal organic frameworks, and carbon bases of nanozymes, transferring electrons to form functional nanozymes[38][39][40][41]. The interaction between functional nanozymes and various interface components was analyzed by generating energy conversion effects (biosensing and signal transduction)[42]. Like target recognition conversion into an optical signal or electrical signal, the probe assembled by gold nanozyme and aptamer AG3 converts the process of specific recognition of murine norovirus into an optical signal[43]; the probe assembled by the gold nanoparticle–graphene oxide hybrid and the respiratory syncytial virus antibody converts the process of specifically recognizing RSV into an optical signal[44]. Like target substance activation conversion into a metabolic signal, Fe3O4 NPs induce AMPK activation and enhance glucose uptake, which has potential effects in diabetes care[45]; organic polymer nanozyme SPNK induces the release of KYNase, which degrades kynurenine (Kyn)[1]. Consequently, it may be true that nanozymes can control immunomodulation. By studying the interface effects of nanozymes in these energy conversion events and analyzing the interactions between various interface components, we further understand functional nanozymes[42].
1.4. Nucleic Acid Modulation for Functional Nanozyme
Since one of the centerpieces of the biochemical energy conversion mechanism is the interface modulation event, more attention has focused on the strategy of interface modulation for target assay. To the best of our knowledge, this article is the first to analyze and summarize such phenomena. This article pays great attention to the interface modulation of nucleic acids to nanozymes: modification, binding, immobilization, and the resulting major changes in interface components in the structure of nanozymes, which lead to an increase or decrease in enzyme catalytic activity[46][47][48][49][50]. The controllability and accuracy of modulation technology is an important force for promoting the progress of social civilization. To cater to the perfect control of enzyme activity in practical applications, scientists have expended great effort in studying the crosstalk between nanozymes and nucleic acids, which retains the advantages of nucleic acids but does not limit the properties of nanozymes that need to be expressed.
2. Crosstalk between Nanozyme and Nucleic Acid
Nucleic acid modulation of nanozymes is a key step in energy conversion events. Studies have shown that nucleic acids can change the size, shape, composition, surface modification, and state of NPs through biological, chemical, and physical effects[51][52][53][54]. Here, we scientifically discuss the construction of three types of crosstalk between nanozymes and nucleic acids: (nanozyme precursor ion)/(nucleic acid) self-assembly, nanozyme-nucleic acid irreversible binding, and NP-nucleic acid reversible binding.
2.1. Self-Assembly Nanozyme
The rich structural features of nucleic acids endow them with diverse binding capabilities with NPs. Due to the noncovalent interactions that occur during the assembly of NPs and nucleic acids, the assembled nanozymes have new interface effects and exhibit the desired morphology, chemical-physical properties, and stimulus responsiveness.
Here, we summarize the interactions among components. Recent studies have confirmed that the presence of various elements commonly found in nuclein—oxygen and nitrogen[55]. Nitrogen acts as an electron donor, and oxygen provides electron pairs, which can hybridize or covalently coordinate with single or multiple metal ion precursors (Figure 1); therefore, they can controllably modulate the enzyme activity and stability of nanoparticles[56][57][58][59][60]. In addition, Chen et al.[59] introduced nanocarrier mesoporous silica based on the study of noncovalent binding and discovered that the nucleic acid acted on the platinum particle precursor ion to produce a “reversible” masking effect at the active site.
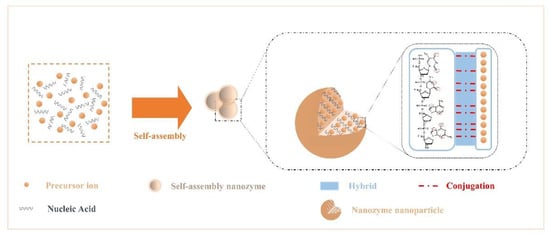
Figure 1. Crosstalk from self-assembled nanozymes: noncovalent interactions that occur during the assembly of NPs and nucleic acids.
2.2. Irreversible Binding Nanozyme
Nucleic acids have a complex skeletal structure composed of bases, phosphate groups, and ribose, so they have a wealth of modification sites[61][62], while NPs have the characteristics of many surface active sites, strong adsorption, and a high density of valence electrons. Researchers have made use of their rich properties, modified them, and accomplished the irreversible combination of the two components. Many modification bridges have been used to promote binding between NPs and nucleic acids (Figure 2), for example, -NH-SiO2-, -biotin-streptavidin-, -NH-COOH-, -C-NH-, -S-, -magnetic bead-, and -NH-[63][64][65][66][67][68][69]. Binding at the interface perfectly merges the advantages of the two. Additionally, irreversible chemical modification may affect an NP’s characteristics and limit the number of connections of nucleic acids. In summary, functional nanozymes have both the enzyme activity of NPs and the target recognition function of nucleic acids, producing the desired interface effect, which can be applied to biochemical energy conversion events.
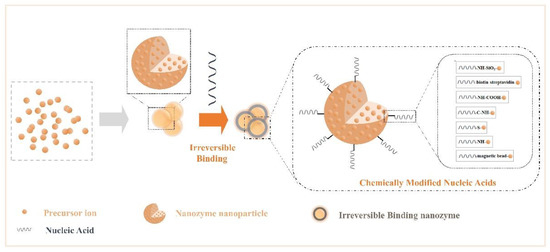
Figure 2. Crosstalk from irreversible binding nanozymes: creating irreversible binding nanozymes based on synthetic materials (such as chemically modified nucleic acids).
2.3. Reversible Binding Nanozyme
Nucleic acids have parallel stacked bases, polymerized anionic phosphate backbones, sugar rings, and active sites composed of large grooves and small grooves formed by double-stranded structures[26], and these features allow nucleic acids to combine/separate with nanozymes with high specific surface areas and rich surface chemical morphologies, as expected in various analytical events based on competitive binding mechanisms[70]. In general, the addition of nucleic acids to the nanozyme reaction system increases the interface components of the NPs, dramatically increases the free energy of the interface with the NPs, and changes the structure-related properties of the interface, such as ionic valence and electronic transfer. Moreover, it can be formed into a reversible binding nanozyme, which preserves the characteristics of nucleic acids and NP as much as possible. Here, we list the general influence of nucleic acid on NPs (Figure 3) as changes in the steric hindrance effect, changes in the number of active sites, changes in the dispersion of NPs, changes in the structure of NPs (such as oxygen vacancies), and changes in substrate activity in producing highly active substances (such as hydroxyl radicals).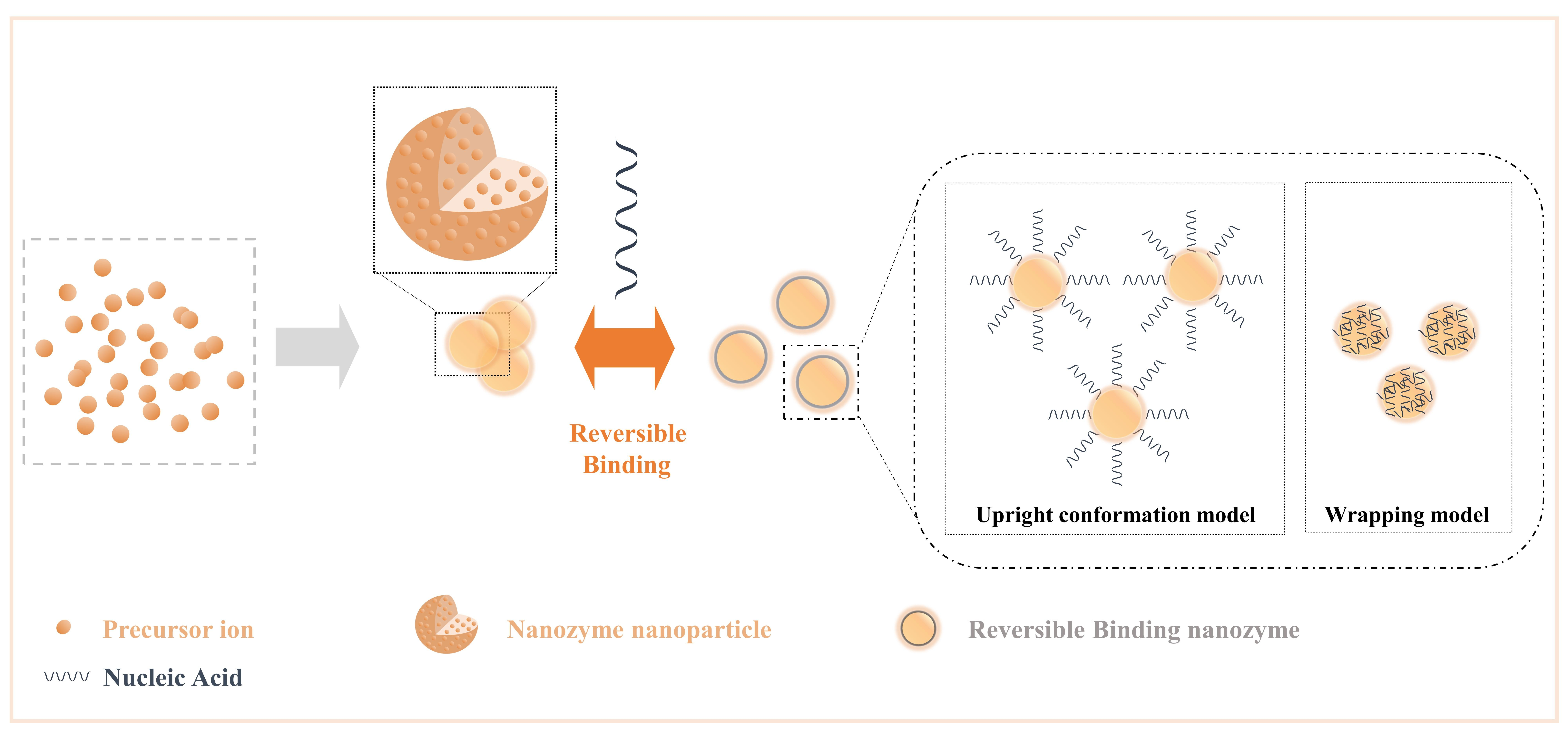
Figure 3. Crosstalk from irreversible binding nanozymes and reversible binding nanozymes allow nucleic acids to combine/separate with nanozymes by the effect of intermolecular forces. Reprinted from reference[70].
This entry is adapted from the peer-reviewed paper 10.3390/catal11050638
References
- Ziling Zeng; Chi Zhang; Jingchao Li; Dong Cui; Yuyan Jiang; Kanyi Pu; Activatable Polymer Nanoenzymes for Photodynamic Immunometabolic Cancer Therapy. Advanced Materials 2020, 33, e2007247, 10.1002/adma.202007247.
- Namrata Singh; Somanathapura K. Naveenkumar; Motika Geethika; Govindasamy Mugesh; A Cerium Vanadate Nanozyme with Specific Superoxide Dismutase Activity Regulates Mitochondrial Function and ATP Synthesis in Neuronal Cells. Angewandte Chemie 2020, 133, 3158-3167, 10.1002/ange.202011711.
- Hui Wei; Erkang Wang; Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes. Chemical Society Reviews 2013, 42, 6060-6093, 10.1039/c3cs35486e.
- Xiaoyu Wang; Yihui Hu; Hui Wei; Nanozymes in bionanotechnology: from sensing to therapeutics and beyond. Inorganic Chemistry Frontiers 2016, 3, 41-60, 10.1039/c5qi00240k.
- Xueqing Xiong; Yanyan Huang; Changxu Lin; Xiang Yang Liu; Youhui Lin; Recent advances in nanoparticulate biomimetic catalysts for combating bacteria and biofilms. Nanoscale 2019, 11, 22206-22215, 10.1039/c9nr05054j.
- Jiangjiexing Wu; Xiaoyu Wang; Quan Wang; Zhangping Lou; Sirong Li; Yunyao Zhu; Li Qin; Hui Wei; Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes (II). Chemical Society Reviews 2018, 48, 1004-1076, 10.1039/c8cs00457a.
- Ronald Breslow; Biomimetic Chemistry and Artificial Enzymes: Catalysis by Design. Accounts of Chemical Research 1995, 28, 146-153, 10.1021/ar00051a008.
- . , , , .
- Yanyan Huang; Jinsong Ren; Xiaogang Qu; Nanozymes: Classification, Catalytic Mechanisms, Activity Regulation, and Applications. Chemical Reviews 2019, 119, 4357-4412, 10.1021/acs.chemrev.8b00672.
- Amit Kumar Dutta; Sudipto Das; Partha Kumar Samanta; Shounak Roy; Bibhutosh Adhikary; Papu Biswas; Non–enzymatic amperometric sensing of hydrogen peroxide at a CuS modified electrode for the determination of urine H2O2. Electrochimica Acta 2014, 144, 282-287, 10.1016/j.electacta.2014.08.051.
- Flavio Manea; Florence Bodar Houillon; Lucia Pasquato; Paolo Scrimin; Nanozymes: Gold-Nanoparticle-Based Transphosphorylation Catalysts. Angewandte Chemie 2004, 116, 6291-6295, 10.1002/ange.200460649.
- Yibo Zhou; Biwu Liu; Ronghua Yang; Juewen Liu; Filling in the Gaps between Nanozymes and Enzymes: Challenges and Opportunities. Bioconjugate Chemistry 2017, 28, 2903-2909, 10.1021/acs.bioconjchem.7b00673.
- Hanjun Cheng; Yufeng Liu; Yihui Hu; Yubin Ding; Shichao Lin; Wen Cao; Qian Wang; Jiangjiexing Wu; Faheem Muhammad; Xiaozhi Zhao; et al. Monitoring of Heparin Activity in Live Rats Using Metal–Organic Framework Nanosheets as Peroxidase Mimics. Analytical Chemistry 2017, 89, 11552-11559, 10.1021/acs.analchem.7b02895.
- Lizeng Gao; Jie Zhuang; Leng Nie; Jinbin Zhang; Yu Zhang; Ning Gu; Taihong Wang; Jing Feng; Dongling Yang; Sarah Perrett; et al. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nature Nanotechnology 2007, 2, 577-583, 10.1038/nnano.2007.260.
- Bing Jiang; Demin Duan; Lizeng Gao; Mengjie Zhou; Kelong Fan; Yan Tang; Juqun Xi; Yuhai Bi; Zhou Tong; George Fu Gao; et al. Standardized assays for determining the catalytic activity and kinetics of peroxidase-like nanozymes. Nature Protocols 2018, 13, 1506-1520, 10.1038/s41596-018-0001-1.
- Youhui Lin; Jinsong Ren; Xiaogang Qu; Catalytically Active Nanomaterials: A Promising Candidate for Artificial Enzymes. Accounts of Chemical Research 2014, 47, 1097-1105, 10.1021/ar400250z.
- Xu Zhu; Lin Tang; Jiajia Wang; Bo Peng; Xilian Ouyang; Jisui Tan; Jiangfang Yu; HaoPeng Feng; Jialin Tang; Enhanced peroxidase-like activity of boron nitride quantum dots anchored porous CeO2 nanorods by aptamer for highly sensitive colorimetric detection of kanamycin. Sensors and Actuators B: Chemical 2021, 330, 129318, 10.1016/j.snb.2020.129318.
- Yuichiro Aiba; Jun Sumaoka; Makoto Komiyama; Artificial DNA cutters for DNA manipulation and genome engineering. Chemical Society Reviews 2011, 40, 5657-5668, 10.1039/c1cs15039a.
- Muthuchamy Maruthupandy; Govindan Rajivgandhi; Thillaichidambaram Muneeswaran; Thirumalaiswamy Vennila; Franck Quero; Ji-Ming Song; Chitosan/silver nanocomposites for colorimetric detection of glucose molecules. International Journal of Biological Macromolecules 2019, 121, 822-828, 10.1016/j.ijbiomac.2018.10.063.
- J. Justin Gooding; Big Moves in Biosensing. ACS Sensors 2016, 1, 633-633, 10.1021/acssensors.6b00362.
- J. Justin Gooding; The Exciting World of Single Molecule Sensors. ACS Sensors 2016, 1, 1163-1164, 10.1021/acssensors.6b00624.
- J. Justin Gooding; Katharina Gaus; Single‐Molecule Sensors: Challenges and Opportunities for Quantitative Analysis. Angewandte Chemie International Edition 2016, 55, 11354-11366, 10.1002/anie.201600495.
- Jiangjiexing Wu; Sirong Li; Hui Wei; Integrated nanozymes: facile preparation and biomedical applications. Chemical Communications 2018, 54, 6520-6530, 10.1039/c8cc01202d.
- Minmin Liang; Xiyun Yan; Nanozymes: From New Concepts, Mechanisms, and Standards to Applications. Accounts of Chemical Research 2019, 52, 2190-2200, 10.1021/acs.accounts.9b00140.
- Leilei Guo; Kaixun Huang; Hongmei Liu; Biocompatibility selenium nanoparticles with an intrinsic oxidase-like activity. Journal of Nanoparticle Research 2016, 18, 1-10, 10.1007/s11051-016-3357-6.
- Fang Pu; Jinsong Ren; Xiaogang Qu; Nucleobases, nucleosides, and nucleotides: versatile biomolecules for generating functional nanomaterials. Chemical Society Reviews 2017, 47, 1285-1306, 10.1039/c7cs00673j.
- Xianzhi Zhang; Yuanchang Liu; Sanjana Gopalakrishnan; Laura Castellanos-Garcia; Gengtan Li; Morgane Malassiné; Imad Uddin; Rui Huang; David C. Luther; Richard W. Vachet; et al. Intracellular Activation of Bioorthogonal Nanozymes through Endosomal Proteolysis of the Protein Corona. ACS Nano 2020, 14, 4767-4773, 10.1021/acsnano.0c00629.
- Yu-Min Wang; Jin-Wen Liu; Gary Adkins; Wen Shen; Michael Patrick Trinh; Lu-Ying Duan; Jian-Hui Jiang; Wenwan Zhong; Enhancement of the Intrinsic Peroxidase-Like Activity of Graphitic Carbon Nitride Nanosheets by ssDNAs and Its Application for Detection of Exosomes. Analytical Chemistry 2017, 89, 12327-12333, 10.1021/acs.analchem.7b03335.
- Xiaowei Li; Yan Zhao; Synthetic glycosidases for the precise hydrolysis of oligosaccharides and polysaccharides. Chemical Science 2020, 12, 374-383, 10.1039/d0sc05338d.
- Di Zhang; Ying-Xi Zhao; Yu-Juan Gao; Fu-Ping Gao; Yun-Shan Fan; Xiao-Jun Li; Zhong-Yu Duan; Hao Wang; Anti-bacterial and in vivo tumor treatment by reactive oxygen species generated by magnetic nanoparticles.. Journal of Materials Chemistry B 2013, 1, 5100-5107, 10.1039/c3tb20907e.
- Jun Ma; Jingjing Qiu; Shiren Wang; Nanozymes for Catalytic Cancer Immunotherapy. ACS Applied Nano Materials 2020, 3, 4925-4943, 10.1021/acsanm.0c00396.
- Taoli Ding; Jing Yang; Victor Pan; Nan Zhao; Zuhong Lu; Yonggang Ke; Cheng Zhang; DNA nanotechnology assisted nanopore-based analysis. Nucleic Acids Research 2020, 48, 2791-2806, 10.1093/nar/gkaa095.
- Sona Sivakova; Stuart J. Rowan; Nucleobases as supramolecular motifs. Chemical Society Reviews 2004, 34, 9-21, 10.1039/b304608g.
- Pei Zhou; Rufei Shi; Jian-Feng Yao; Chuan-Fang Sheng; Hui Li; Supramolecular self-assembly of nucleotide–metal coordination complexes: From simple molecules to nanomaterials. Coordination Chemistry Reviews 2015, 292, 107-143, 10.1016/j.ccr.2015.02.007.
- Sandeep Verma; Ashutosh Kumar Mishra; Jitendra Kumar; The Many Facets of Adenine: Coordination, Crystal Patterns, and Catalysis. Accounts of Chemical Research 2009, 43, 79-91, 10.1021/ar9001334.
- Artur Ciesielski; Mohamed El Garah; Stefano Masiero; Paolo Samorì; Self-assembly of Natural and Unnatural Nucleobases at Surfaces and Interfaces. Small 2015, 12, 83-95, 10.1002/smll.201501017.
- Gretchen Marie Peters; Jeffery T. Davis; Supramolecular gels made from nucleobase, nucleoside and nucleotide analogs. Chemical Society Reviews 2016, 45, 3188-3206, 10.1039/c6cs00183a.
- Zuccheri, Giampaolo; Samorì, Bruno; DNA Nanotechnology. Advanced Structural Safety Studies 2011, 749, 1-361, 10.1007/978-1-61779-142-0.
- Chun-Hua Lu; Alessandro Cecconello; Itamar Willner; Recent Advances in the Synthesis and Functions of Reconfigurable Interlocked DNA Nanostructures. Journal of the American Chemical Society 2016, 138, 5172-5185, 10.1021/jacs.6b00694.
- Katsuyuki Aoki; Kazutaka Murayama; Nucleic Acid-Metal Ion Interactions in the Solid State. Metal Ions in Life Sciences 2011, 10, 43-102, 10.1007/978-94-007-2172-2_2.
- Helmut Sigel; Rolf Griesser; Nucleoside 5′-triphosphates: self-association, acid–base, and metal ion-binding properties in solution. Chemical Society Reviews 2005, 34, 875-900, 10.1039/b505986k.
- Xiaomei Shen; Wenqi Liu; Xuejiao Gao; Zhanghui Lu; Xiaochun Wu; Xingfa Gao; Mechanisms of Oxidase and Superoxide Dismutation-like Activities of Gold, Silver, Platinum, and Palladium, and Their Alloys: A General Way to the Activation of Molecular Oxygen. Journal of the American Chemical Society 2015, 137, 15882-15891, 10.1021/jacs.5b10346.
- Pabudi Weerathunge; Rajesh Ramanathan; Valeria A. Torok; Kate Hodgson; Yun Xu; Royston Goodacre; Bijay Kumar Behera; Vipul Bansal; Ultrasensitive Colorimetric Detection of Murine Norovirus Using NanoZyme Aptasensor. Analytical Chemistry 2019, 91, 3270-3276, 10.1021/acs.analchem.8b03300.
- Lei Zhan; Chun Mei Li; Wen Bi Wu; Cheng Zhi Huang; A colorimetric immunoassay for respiratory syncytial virus detection based on gold nanoparticles–graphene oxide hybrids with mercury-enhanced peroxidase-like activity. Chemical Communications 2014, 50, 11526-11528, 10.1039/c4cc05155f.
- Yanfeng Zhou; Chang Liu; Yun Yu; Min Yin; Jinli Sun; Jing Huang; Nan Chen; Hui Wang; Chunhai Fan; Haiyun Song; et al. An Organelle‐Specific Nanozyme for Diabetes Care in Genetically or Diet‐Induced Models. Advanced Materials 2020, 32, e2003708, 10.1002/adma.202003708.
- Peng Gao; Xin Chang; Dagan Zhang; Yafei Cai; Gen Chen; Hao Wang; Tianfu Wang; Synergistic integration of metal nanoclusters and biomolecules as hybrid systems for therapeutic applications. Acta Pharmaceutica Sinica B 2021, 11, 1175-1199, 10.1016/j.apsb.2020.12.004.
- Ming Wei; Yanxia Qiao; Haitao Zhao; Jie Liang; Ting Shuai Li; Yongsong Luo; Siyu Lu; Xifeng Shi; Wenbo Lu; Xuping Sun; et al. Electrochemical non-enzymatic glucose sensors: recent progress and perspectives. Chemical Communications 2020, 56, 14553-14569, 10.1039/d0cc05650b.
- Nai‐Chang Lo; Wei‐Shan Hsu; Yi‐Ting Chen; I‐Wen Sun; Po‐Yu Chen; Facile Nonenzymatic Glucose Electrode Composed of Commercial CuO Powder and Ionic Liquid Binder. Electroanalysis 2020, 33, 909-915, 10.1002/elan.202060467.
- Xiaomin Qian; Isabella Nymann Westensee; Edit Brodszkij; Brigitte Städler; Cell mimicry as a bottom‐up strategy for hierarchical engineering of nature‐inspired entities. WIREs Nanomedicine and Nanobiotechnology 2020, 13, e1683, 10.1002/wnan.1683.
- Nirmal Goswami; KaiYuan Zheng; Jianping Xie; Bio-NCs – the marriage of ultrasmall metal nanoclusters with biomolecules. Nanoscale 2014, 6, 13328-13347, 10.1039/c4nr04561k.
- Xiaoqi Tao; Xin Wang; Biwu Liu; Juewen Liu; Conjugation of antibodies and aptamers on nanozymes for developing biosensors. Biosensors and Bioelectronics 2020, 168, 112537, 10.1016/j.bios.2020.112537.
- Li Shang; Shaojun Dong; Gerd Ulrich Nienhaus; Ultra-small fluorescent metal nanoclusters: Synthesis and biological applications. Nano Today 2011, 6, 401-418, 10.1016/j.nantod.2011.06.004.
- Shin-Ichi Tanaka; Jun Miyazaki; Dhermendra Tiwari; Takashi Jin; Yasushi Inouye; Fluorescent Platinum Nanoclusters: Synthesis, Purification, Characterization, and Application to Bioimaging. Angewandte Chemie International Edition 2010, 50, 431-435, 10.1002/anie.201004907.
- Hideya Kawasaki; Hiroko Yamamoto; Hiroki Fujimori; Ryuichi Arakawa; Mitsuru Inada; Yasuhiko Iwasaki; Surfactant-free solution synthesis of fluorescent platinum subnanoclusters. Chemical Communications 2010, 46, 3759-3761, 10.1039/b925117k.
- Ralf Dahm; Friedrich Miescher and the discovery of DNA. Developmental Biology 2005, 278, 274-288, 10.1016/j.ydbio.2004.11.028.
- Yan Fu; Xuyin Zhao; Jinli Zhang; Wei Li; DNA-Based Platinum Nanozymes for Peroxidase Mimetics. The Journal of Physical Chemistry C 2014, 118, 18116-18125, 10.1021/jp503242e.
- Yanhua Sun; Jun Wang; Wei Li; Jinli Zhang; Yaodan Zhang; Yan Fu; DNA-stabilized bimetallic nanozyme and its application on colorimetric assay of biothiols. Biosensors and Bioelectronics 2015, 74, 1038-1046, 10.1016/j.bios.2015.08.001.
- Weiwei Chen; Xueen Fang; Hua Li; Hongmei Cao; Jilie Kong; DNA-mediated inhibition of peroxidase-like activities on platinum nanoparticles for simple and rapid colorimetric detection of nucleic acids. Biosensors and Bioelectronics 2017, 94, 169-175, 10.1016/j.bios.2017.02.025.
- Weiwei Chen; Xueen Fang; Xin Ye; Xinjun Wang; Jilie Kong; Colorimetric DNA assay by exploiting the DNA-controlled peroxidase mimicking activity of mesoporous silica loaded with platinum nanoparticles. Microchimica Acta 2018, 185, 544, 10.1007/s00604-018-3026-9.
- Akon Higuchi; Yi-Di Siao; Siou-Ting Yang; Pei-Vin Hsieh; Hisashi Fukushima; Yung Chang; Ruoh-Chyu Ruaan; Wen-Yih Chen; Preparation of a DNA Aptamer−Pt Complex and Its Use in the Colorimetric Sensing of Thrombin and Anti-Thrombin Antibodies. Analytical Chemistry 2008, 80, 6580-6586, 10.1021/ac8006957.
- C. V. Kumar; Emma H. Asuncion; DNA binding studies and site selective fluorescence sensitization of an anthryl probe. Journal of the American Chemical Society 1993, 115, 8547-8553, 10.1021/ja00072a004.
- Seyed Zachariah Moradi; Amin Nowroozi; Komail Sadrjavadi; Sajad Moradi; Kamran Mansouri; Leila Hosseinzadeh; Mohsen Shahlaei; Direct evidences for the groove binding of the Clomifene to double stranded DNA. International Journal of Biological Macromolecules 2018, 114, 40-53, 10.1016/j.ijbiomac.2018.03.040.
- Peng Hu; Lei Han; Chengzhou Zhu; Shao Jun Dong; Nanoreactors: a novel biosensing platform for protein assay. Chemical Communications 2013, 49, 1705-1707, 10.1039/c2cc37734a.
- Ling Zhang; Zhengnan Qi; Yan Zou; Jiaxing Zhang; Wenjun Xia; Rui Zhang; Zhiyan He; Xiaoxiao Cai; Yunfeng Lin; Sheng-Zhong Duan; et al. Engineering DNA–Nanozyme Interfaces for Rapid Detection of Dental Bacteria. ACS Applied Materials & Interfaces 2019, 11, 30640-30647, 10.1021/acsami.9b10718.
- Lisha Zhang; Ru Huang; Weipeng Liu; Hongxing Liu; XiaoMing Zhou; Da Xing; Rapid and visual detection of Listeria monocytogenes based on nanoparticle cluster catalyzed signal amplification. Biosensors and Bioelectronics 2016, 86, 1-7, 10.1016/j.bios.2016.05.100.
- Zhanxia Zhang; Zhongjun Wang; Xiaolei Wang; Xiurong Yang; Magnetic nanoparticle-linked colorimetric aptasensor for the detection of thrombin. Sensors and Actuators B: Chemical 2010, 147, 428-433, 10.1016/j.snb.2010.02.013.
- Duanping Sun; Xiangan Lin; Jing Lu; Ping Wei; Zibin Luo; Xiange Lu; Zuanguang Chen; Luyong Zhang; DNA nanotetrahedron-assisted electrochemical aptasensor for cardiac troponin I detection based on the co-catalysis of hybrid nanozyme, natural enzyme and artificial DNAzyme. Biosensors and Bioelectronics 2019, 142, 111578, 10.1016/j.bios.2019.111578.
- Zahra Dehghani; Trieu Nguyen; Mohsen Golabi; Morteza Hosseini; Ali H. Rezayan; Javad Mohammadnejad; Anders Wolff; Aaydha C. Vinayaka; Magnetic beads modified with Pt/Pd nanoparticle and aptamer as a catalytic nano-bioprobe in combination with loop mediated isothermal amplification for the on-site detection of Salmonella Typhimurium in food and fecal samples. Food Control 2021, 121, 107664, 10.1016/j.foodcont.2020.107664.
- YongMei Wu; Gaiping Li; Lina Zou; Sheng Lei; Qian Yu; Baoxian Ye; Highly active DNAzyme-peptide hybrid structure coupled porous palladium for high-performance electrochemical aptasensing platform. Sensors and Actuators B: Chemical 2018, 259, 372-379, 10.1016/j.snb.2017.12.091.
- Jing An; Galong Li; Yifan Zhang; Tingbin Zhang; Xiaoli Liu; Fei Gao; Mingli Peng; Yuan He; Haiming Fan; Recent Advances in Enzyme-Nanostructure Biocatalysts with Enhanced Activity. Catalysts 2020, 10, 338, 10.3390/catal10030338.
