Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Neurosciences
Neurodegenerative diseases (NDs) include a large spectrum of heterogeneous and multifactorial pathologies, such as amyotrophic lateral sclerosis, frontotemporal dementia, Alzheimer’s disease, Parkinson’s disease, Huntington’s disease and multiple system atrophy, and the formation of inclusion bodies resulting from protein misfolding and aggregation is a hallmark of these disorders.
- neurodegenerative disease
- RNA-binding protein
1. Introduction
Neurodegenerative diseases (NDs) are progressive and fatal diseases characterized by selective neuronal vulnerability with degeneration in specific brain regions, and deposits of abnormal proteins are often found in the brains of ND patients [1]. NDs are age-dependent disorders and have recently become a major medical and social threat, in part due to the growing population of elderly people in super-aged societies worldwide. These disorders include Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD) and multiple system atrophy (MSA). Their symptoms partly overlap, with some causing memory and cognitive impairments and others affecting a person’s ability to move, speak and breathe. NDs impose a major burden on societies, families and individuals, not only through the loss of labor but, also, through the frequent requirement of 24-h support by the relatives of the affected individual. The development of therapeutics effective for NDs is absolutely crucial to solving this problem, and to this end, it is necessary to understand the causes and mechanisms of each disease.
2. Involvement of RNA-Binding Proteins in Neuronal microRNA Biogenesis
Increasing lines of evidence suggests that functional interactions and cooperation between RBPs and miRNAs are a key mechanism in the post-transcriptional regulation of gene expression [101]. miRNAs are a class of short noncoding RNAs that are approximately twenty nucleotides in length [11]. Their function consists mostly of silencing target expressions by binding to target gene transcripts located mainly at the 3’-UTR. Most miRNA genes are located in intergenic regions or in an antisense orientation to gene regions on the genome. Clustered miRNAs can either be simultaneously transcribed from single polycistronic transcripts containing multiple miRNAs or independently transcribed. The biogenesis of miRNAs is precisely regulated by molecular complexes including RBPs at almost every step [102] (Figure 1). The primary miRNAs (pri-miRNAs)—that are primary transcripts containing stem-loop structures and are usually thousands of nucleotides in length—are initially transcribed by a polymerase—in most cases, RNA polymerase II. Then, the pri-miRNAs are cleaved by a complex called a Microprocessor, which contains the ribonuclease III Drosha and the RNA-binding protein DGCR8/Pasha. The Microprocessor generates small hairpin-shaped RNAs of approximately 70–100 nucleotides in length called miRNA precursors (pre-miRNAs). This step has been reported to involve NDs-associated RBPs such as TDP-43, FET proteins and hnRNPs. TDP-43 associates with the Drosha complex and binds to a set of pri-miRNAs, such as pri-miR-132, -143, -558 and -574, and promotes their cleavage [103]. FUS interacts with specific neuronal pri-miRNA sequences, together with Drosha, and assists in recruiting Drosha to the chromatin, allowing the efficient processing of neuronal miRNAs such as pri-miR-9, -125b and -132 [104]. In addition, the specific and prevalent interactions between EWS and Drosha with a large subset of pri-miRNAs such as pri-miR-29b, -18b, -34a, -222 and -let7 were shown to enhance the recruitment of Drosha to chromatin [105,106]. The HuR-mediated binding of Msi2 to the conserved terminal loop of pri-miR-7 also inhibits Drosha cleavage [107]. Moreover, hnRNP A1 binding to pri-let-7 leads to the inhibition of Drosha processing, whereas binding to pri-miR-18a contributes to the stimulation of processing [108,109,110,111]. In the next step, pre-miRNAs exported by exportin-5 in complex with RAN-GTP are processed by a double-stranded ribonuclease III enzyme, termed Dicer, which is complexed with a double-stranded RNA-binding protein, TRBP. AUF1 has a role in suppressing miRNA production by reducing Dicer production in association with the endogenous Dicer mRNA [112]. TDP-43 is also involved in this step by facilitating processing through the binding of Dicer to specific sites of pre-miRNAs such as pre-miR-143 and -574 [103]. Next, an Argonaute protein forms a part of an effector complex, called the RNA-induced silencing complex (RISC), onto which the mature miRNA duplexes are loaded. TDP-43 is negatively involved in the step of loading specific miRNAs such as miR-1 and -206 onto the RISC complex [113]. Conversely, the complex formed by the association of PABP with mRNA promotes miRISC recruitment [114]. Finally, one strand of the miRNA is removed from RISC to generate the mature RISC that induces gene silencing. The post-transcriptional regulation by the RISC complex is mediated by the incomplete base pairing of miRNA–mRNA interactions, likely due to the targeting of multiple transcripts, which contributes to the complexity or redundancy of miRNA systems.
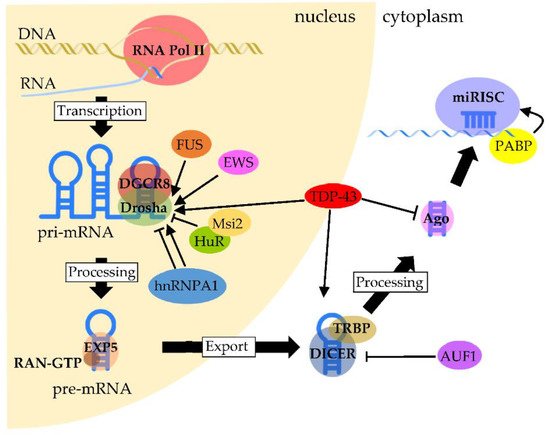
Figure 1. Regulation of RBPs in miRNA biogenesis and processing. Pri-miRNAs are transcribed mainly by RNA polymerase II. The processing of pri-miRNA is mediated by a complex formed between Drosha and DGCR8, called Microprocessor, to generate pre-miRNA. TDP-43 facilitates the association of the Drosha complex with pri-miRNAs, whereas FUS and EWS promote Drosha recruitment to chromatin. HuR-mediated Msi2 binding to pri-miRNA inhibits Drosha cleavage. The function of hnRNP A1 is complexed, since it contributes to either the inhibition or stimulation of processing, which depends on the pri-miRNA. After pre-miRNAs are exported through the binding of Exportin-5 with RAN-GTP, pre-miRNAs are processed by Dicer. The processing of pre-miRNAs is facilitated by TDP-43 through the binding of Dicer with pre-miRNAs, whereas Dicer expression is negatively regulated by AUF1. On the other hand, TDP-43 negatively regulates the formation of miRISC from the Ago protein. PABP binds to the poly(A) region of target mRNA and promotes miRISC recruitment.
3. Interplay between RNA-Binding Proteins and microRNAs in Neurodegenerative Disease
The progressive degeneration of the structure and function of the nervous system is the main feature of NDs such as AD, PD, ALS, FTD, HD and MSA [1]. These diseases primarily occur in the later stages of life. As an ironic side effect of the worldwide increases in life expectancy and resulting growth in the aging population, the number of people suffering from NDs has also grown. Currently, no NDs are curable; the available treatments only manage the symptoms or halt the disease progression.
The genetic mutations that cause the pathogenesis of familial NDs have been well-studied; however, with the exception of HD, the majority of NDs are sporadic. Our knowledge of sporadic NDs is unfortunately not sufficient to understand their molecular etiologies, even though their pathologies are well-established. Several lines of evidence indicate that the onset of sporadic diseases is due to the synergistic effect of genetic polymorphism, environmental factors or their combination with overlapping disease mechanisms, as seen in the familial disease pathologies [115]. In fact, gene expression profiling from clinical studies has revealed a profound dysregulation of gene expression in the tissues of patients with NDs [116], suggesting that the post-transcriptional and/or post-translational regulation of the gene expression would clearly affect the onset or progression of NDs.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22105292
This entry is offline, you can click here to edit this entry!
