Cathepsin K is a papain-like cysteine protease with high matrix-degrading activity. Among several cathepsins, cathepsin K is the most potent mammalian collagenase, mainly expressed by osteoclasts. This review summarizes most of the recent findings of cathepsin K expression, highlighting its role in renal tumors for diagnostic purposes and as a potential molecular target. Indeed, cathepsin K is a recognized diagnostic tool for the identification of TFE3/TFEB-rearranged renal cell carcinoma, TFEB-amplified renal cell carcinoma, and pure epithelioid PEComa/epithelioid angiomyolipoma. More recently, its expression has been observed in a subgroup of eosinophilic renal neoplasms molecularly characterized by TSC/mTOR gene mutations. Interestingly, both TSC mutations or TFE3 rearrangement have been reported in pure epithelioid PEComa/epithelioid angiomyolipoma. Cathepsin K seems to be a downstream marker of TFE3/TFEB rearrangement, TFEB amplification, and mTOR pathway activation. Given the established role of mTOR inhibitors as a pharmacological option in renal cancers, cathepsin K could be of use as a predictive marker of therapy response and as a potential target. In the future, uropathologists may implement the use of cathepsin K to establish a diagnosis among renal tumors with clear cells, papillary architecture, and oncocytic features.
- cathepsin K
- renal cancers
- PEComa
- translocation renal cell carcinoma
- differential diagnosis
- predictive markers
- TSC1/TSC2
- mTOR pathway
- angiomyolipoma
1. Introduction
| Histotype | Morphological Features | Molecular Alteration | Cathepsin K | HMB45/Melan-A | PAX8 | CD68(PG-M1) |
|---|---|---|---|---|---|---|
| TFE3-rearranged RCC | clear cells in nests | ASPCR1-TFE3 fusion | negative | variable | positive | negative |
| papillary architecture | PRCC-TFE3 fusion | positive | variable | positive | negative | |
| variable | SFPQ-TFE3 fusion | variable | variable | positive | negative | |
| TFEB-rearranged RCC | biphasic appearance | MALAT1-TFEB fusion | positive | positive | positive | negative |
| TFEB-amplified RCC | high grade | TFEB amplification | positive | positive | positive | negative |
| PEComa | epithelioid cells | TSC2 mutation | positive | positive | negative | positive |
| epithelioid cells | SFPQ-TFE3 fusion | positive | positive | negative | positive | |
| ESC-RCC | eosinophilic solid and cystic | TSC1/TSC2 mutation | positive | negative | positive | n.a. |
| EVT | high grade oncocytic | TSC2/mTOR mutation | positive | negative | positive | n.a. |
2. Cathepsin K in the Differential Diagnosis
| Pattern | Histotype | Cathepsin K |
|---|---|---|
| clear cell | clear cell RCC | negative |
| clear cell papillary RCC | negative | |
| chromophobe RCC | negative | |
| TFE3-rearranged RCC | variable | |
| TFEB-rearranged RCC | positive | |
| PEComa | positive | |
| papillary architecture | papillary RCC | negative |
| clear cell papillary RCC | negative | |
| TFE3-rearranged RCC | variable | |
| TFEB-rearranged RCC | positive | |
| oncocytic cells | oncocytoma | negative |
| chromophobe RCC | negative | |
| TFE3-rearranged RCC | variable | |
| TFEB-rearranged RCC | positive | |
| PEComa | positive | |
| ESC-RCC | positive | |
| eosinophilic vacuolated tumor | positive |
3. Conclusions
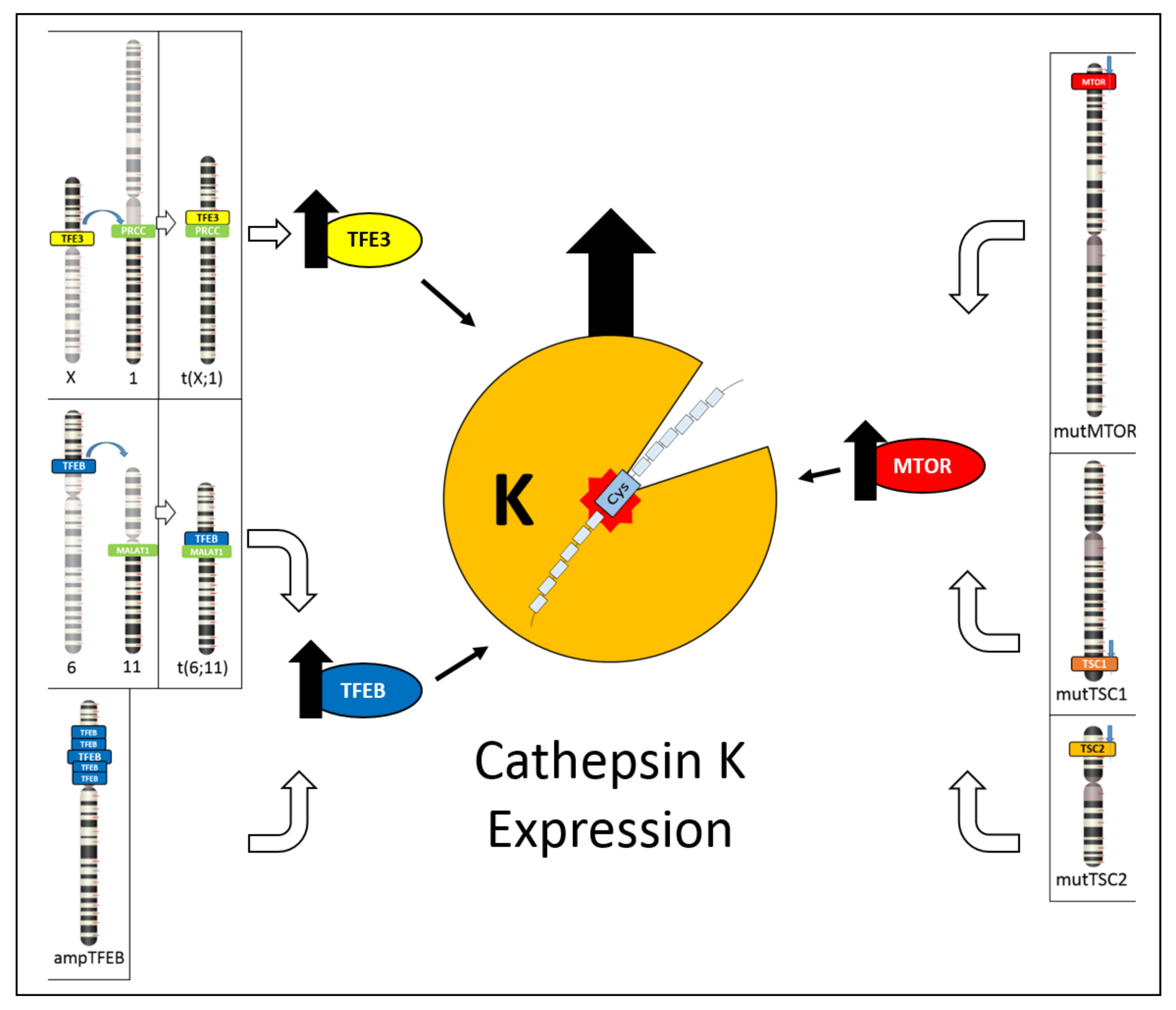 Figure 6. Schematic illustration showing the different mechanisms leading to cathepsin K expression. On the left, TFE3 hyperexpression due to TFE3 gene translocation and TFEB hyperexpression due to either TFEB gene translocation or TFEB gene amplification cause cathepsin K expression. On the right, inactivating mutations of TSC1/TSC2 genes or activating mutations of mTOR gene causes mTOR pathway activation, resulting in cathepsin K expression.
Figure 6. Schematic illustration showing the different mechanisms leading to cathepsin K expression. On the left, TFE3 hyperexpression due to TFE3 gene translocation and TFEB hyperexpression due to either TFEB gene translocation or TFEB gene amplification cause cathepsin K expression. On the right, inactivating mutations of TSC1/TSC2 genes or activating mutations of mTOR gene causes mTOR pathway activation, resulting in cathepsin K expression.This entry is adapted from the peer-reviewed paper 10.3390/cancers13102441
References
- McGrath, M.E.; Klaus, J.L.; Barnes, M.G.; Bromme, D. Crystal structure of human cathepsin K complexed with a potent inhibitor. Nat. Struct. Biol. 1997, 4, 105–109.
- Novinec, M.; Lenarcic, B. Cathepsin K: A unique collagenolytic cysteine peptidase. Biol. Chem. 2013, 394, 1163–1179.
- Costa, A.G.; Cusano, N.E.; Silva, B.C.; Cremers, S.; Bilezikian, J.P. Cathepsin K: Its skeletal actions and role as a therapeutic target in osteoporosis. Nat. Rev. Rheumatol. 2011, 7, 447–456.
- Garber, K. Two pioneering osteoporosis drugs finally approach approval. Nat. Rev. Drug Discov. 2016, 15, 445–446.
- Troen, B.R. The regulation of cathepsin K gene expression. Ann. N. Y. Acad. Sci. 2006, 1068, 165–172.
- Dai, R.; Wu, Z.; Chu, H.Y.; Lu, J.; Lyu, A.; Liu, J.; Zhang, G. Cathepsin K: The action in and beyond bone. Front. Cell. Dev. Biol. 2020, 8, 433.
- Buhling, F.; Rocken, C.; Brasch, F.; Hartig, R.; Yasuda, Y.; Saftig, P.; Bromme, D.; Welte, T. Pivotal role of cathepsin K in lung fibrosis. Am. J. Pathol. 2004, 164, 2203–2216.
- Runger, T.M.; Quintanilla-Dieck, M.J.; Bhawan, J. Role of cathepsin K in the turnover of the dermal extracellular matrix during scar formation. J. Investig. Dermatol. 2007, 127, 293–297.
- Verbovsek, U.; van Noorden, C.J.; Lah, T.T. Complexity of cancer protease biology: Cathepsin K expression and function in cancer progression. Semin. Cancer Biol. 2015, 35, 71–84.
- Reuter, V.E.; Argani, P.; Zhou, M.; Delahunt, B. Members of the IIiDUPG. Best practices recommendations in the application of immunohistochemistry in the kidney tumors: Report from the International Society of Urologic Pathology consensus conference. Am. J. Surg. Pathol. 2014, 38, e35–e49.
- Zheng, G.; Martignoni, G.; Antonescu, C.; Montgomery, E.; Eberhart, C.; Netto, G.; Taube, J.; Westra, W.; Epstein, J.I.; Lotan, T.; et al. A broad survey of cathepsin K immunoreactivity in human neoplasms. Am. J. Clin. Pathol. 2013, 139, 151–159.
- Gupta, S.; Argani, P.; Jungbluth, A.A.; Chen, Y.B.; Tickoo, S.K.; Fine, S.W.; Gopalan, A.; Al-Ahmadie, H.A.; Sirintrapun, S.J.; Sanchez, A.; et al. TFEB expression profiling in renal cell carcinomas: Clinicopathologic correlations. Am. J. Surg. Pathol. 2019, 43, 1445–1461.
- Ross, H.; Martignoni, G.; Argani, P. Renal Cell Carcinoma With Clear Cell and Papillary Features. Arch. Pathol. Lab. Med. 2012, 136, 391–399.
- Caliò, A.; Brunelli, M.; Segala, D.; Pedron, S.; Remo, A.; Ammendola, S.; Munari, E.; Pierconti, F.; Mosca, A.; Bollito, E.; et al. Comprehensive analysis of 34 MiT family translocation renal cell carcinomas and review of the literature: Investigating prognostic markers and therapy targets. Pathology 2020, 52, 297–309.
- Caliò, A.; Segala, D.; Munari, E.; Brunelli, M.; Martignoni, G. MiT Family Translocation Renal Cell Carcinoma: From the Early Descriptions to the Current Knowledge. Cancers 2019, 11, 1110.
- Caliò, A.; Brunelli, M.; Segala, D.; Pedron, S.; Tardanico, R.; Remo, A.; Gobbo, S.; Meneghelli, E.; Doglioni, C.; Hes, O.; et al. t(6;11) renal cell carcinoma: A study of seven cases including two with aggressive behavior, and utility of CD68 (PG-M1) in the differential diagnosis with pure epithelioid PEComa/epithelioid angiomyolipoma. Mod. Pathol. 2018, 31, 474–487.
- Iakymenko, O.A.; Delma, K.S.; Jorda, M.; Kryvenko, O.N. Cathepsin K (Clone EPR19992) Demonstrates Uniformly Positive Immunoreactivity in Renal Oncocytoma, Chromophobe Renal Cell Carcinoma, and Distal Tubules. Int. J. Surg. Pathol. 2021.
- Martignoni, G.; Bonetti, F.; Chilosi, M.; Brunelli, M.; Segala, D.; Amin, M.B.; Argani, P.; Eble, J.N.; Gobbo, S.; Pea, M. Cathepsin K expression in the spectrum of perivascular epithelioid cell (PEC) lesions of the kidney. Mod. Pathol. 2011, 25, 100–111.
- Caron, A.; Richard, D.; Laplante, M. The Roles of mTOR Complexes in Lipid Metabolism. Annu. Rev. Nutr. 2015, 35, 321–348.
- Kim, L.C.; Cook, R.S.; Chen, J. mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene 2017, 36, 2191–2201.
- Han, J.; Wei, L.; Xu, W.; Lu, J.; Wang, C.; Bao, Y.; Jia, W. CTSK inhibitor exert its anti-obesity effects through regulating adipocyte differentiation in high-fat diet induced obese mice. Endocr. J. 2015, 62, 309–317.
