Non-coding RNAs (ncRNAs) are involved in the regulation of cell metabolism and neoplastic transformation. Recent studies have tried to clarify the significance of these information carriers in the genesis and progression of various cancers and their use as biomarkers for the disease; possible targets for the inhibition of growth and invasion by the neoplastic cells have been suggested. The significance of ncRNAs in lung cancer, bladder cancer, kidney cancer, and melanoma has been amply investigated with important results. Recently, the role of long non-coding RNAs (lncRNAs) has also been included in cancer studies. Studies on the relation between endometrial cancer (EC) and ncRNAs, such as small ncRNAs or micro RNAs (miRNAs), transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), antisense RNAs (asRNAs), small nuclear RNAs (snRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), competing endogenous RNAs (ceRNAs), lncRNAs, and long intergenic ncRNAs (lincRNAs) have been published. The recent literature produced in the last three years was extracted from PubMed by two independent readers, which was then selected for the possible relation between ncRNAs, oncogenesis in general, and EC in particular.
- endometrial cancer
- epigenetics
- non-coding RNA (ncRNA)
- long non-coding RNA (lncRNA)
- small non-coding RNA (small ncRNA)
1. Introduction
2. Long Non-Coding RNAs: General Introduction
3. Competing Endogenous RNAs, miRNA, lncRNA Profiling Regulation and Cancer
4. Regulation of Cell Growth by Long Non-Coding RNAs in Endometrioid Endometrial Adenocarcinoma
5. Long Intergenic Non-Protein Coding RNA, Regulator of Reprogramming and Endometrioid Endometrial Adenocarcinoma
6. Micro IRNA in Endometrioid Endometrial Adenocarcinoma
Micro RNA as a Marker and Promoter of Tumor Aggression in EEC
7. Conclusions
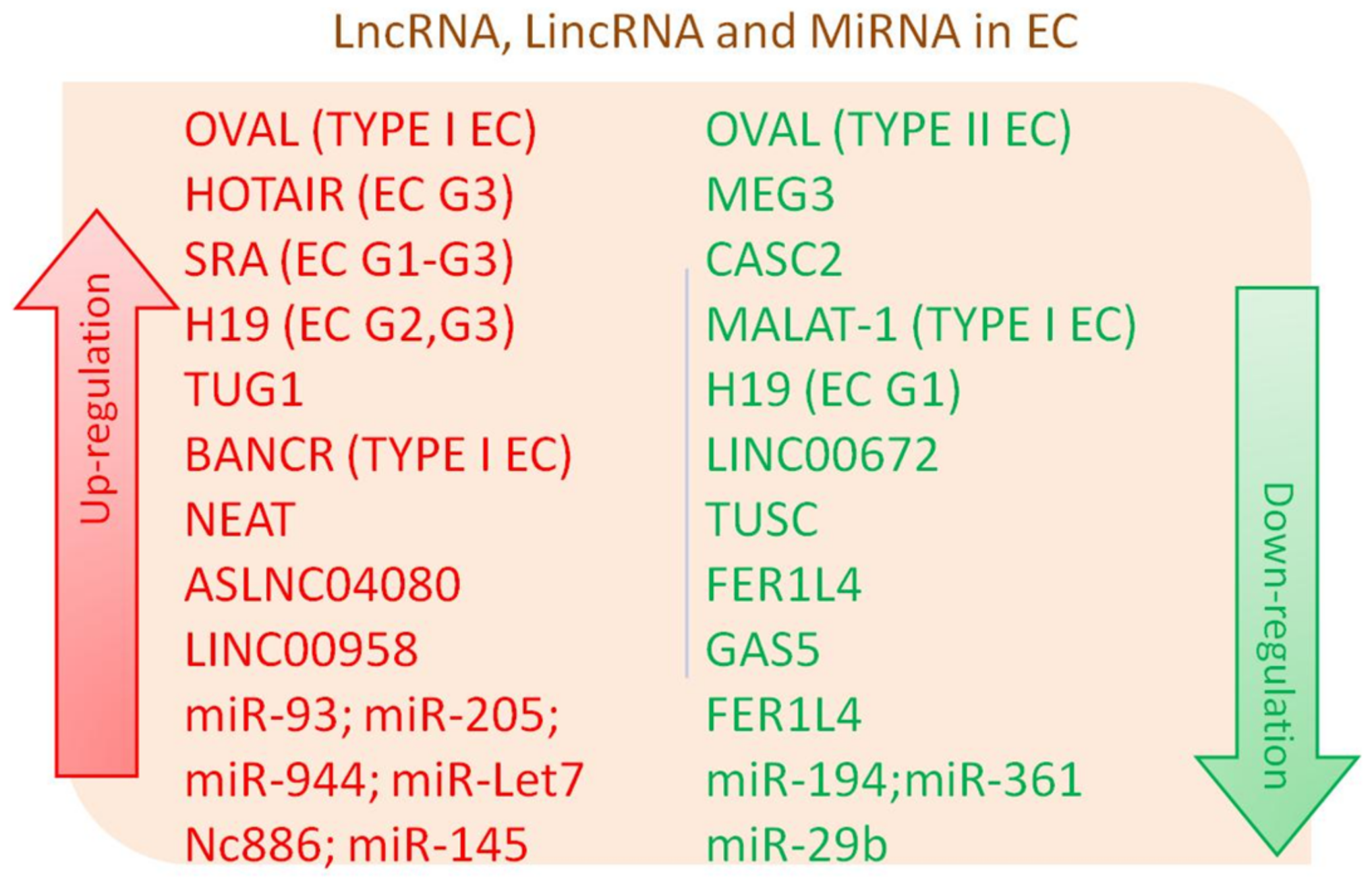 Figure 1. Differentially expressed long non-coding RNA (lncRNA), long intergenic non-coding RNAs (lincRNAs) lincRNA and micro RNAs (miRNA) in Endometrial Cancer (EC).
Figure 1. Differentially expressed long non-coding RNA (lncRNA), long intergenic non-coding RNAs (lincRNAs) lincRNA and micro RNAs (miRNA) in Endometrial Cancer (EC).| NcRNA | Type of ncRNA Regulation/Interaction | Type of EC Regulation/Pathway Interaction | In Vivo or In Vitro Assay or Humans | Reference |
|---|---|---|---|---|
| LncRNA OVAL | down and upregulation | downregulation in type II EC and upregulation in type I EC | In vitro assay from EC tissue | [6] |
| LncRNA MALAT-1 | downregulation/interaction with miR-200 | downregulation in EC through miR-200 | In vitro assay from EC tissue, xenograft tumor model | [14] |
| LncRNA TUG1 | interaction with miR-299/miR-34a-5p | promotion of EC via miR-299 and miR-34a-5p inhibition | In vivo mouse assay, in vitro assay from EC tissue and HEC-1-A cell lines | [29] |
| LncRNAs HOTAIR | upregulation | downregulation in Cisplatin-Resistant Ishikawa Cells (CRIC) and parental IC treated with cisplatin; up-regulation with EC tumor grade increase | In vitro assay CRIC cells; in vitro assay from EC tissue | [19][23] |
| LncRNAs H19 | upregulation | upregulation with EC tumor grade increase and other features associated with poor prognosis | In vitro assay from EC tissues and EC cell line | [35] |
| LncRNAs SRA | upregulation | upregulation regardless of histological tumor grade | In vitro assay from EC tissues | [25] |
| LncRNA BANCR | upregulation | promotion of EE cell proliferation and invasion by MMP2 and MMP1 regulation via ERK–MAPK signaling pathway in type 1 EC | In vitro assay from EC tissues | [37] |
| LncRNAs panel (e.g., KIAA0087, RP11-501O2, FAM212B-AS1, LOC102723552, RP11-140I24 and RP11-600K151) | differential regulation/interaction with 188 mRNAs | differential expression in type I EC | In vitro assay from EC tissues | [27] |
| LncRNA CASC2 | downregulation | potential role as a tumor suppressor | In vitro assay from EC tissues | [18] |
| LncRNA ASLNC04080 | upregulation | upregulation in 22–24 EC tissues and HEC-1-B cell line | In vitro assay from EC tissues and EC cell line | [32] |
| LncRNAs panel (e.g., FLJ27354, RP11-275I14.4, VIM-AS1, CTB-51J22.1 and RP11-229P13.20) | up and downregulation | potential biomarkers of Uterine Corpus (UCEC) | In vitro assay from EC tissues | [34] |
| LncRNA TUSC7 | downregulation | related to EC tumorigenesis and progression | In vitro assay from EC tissues | [39] |
| LncRNA FER1L4 | downregulation | decrease of cell proliferation in FER1L4-overexpressing cells | In vitro assay from EC cell line | [41] |
| LncRNA NEAT | upregulation | increase of Nuclear Enriched Abundant Transcript 1 (NEAT1) in EC | In vitro assay from EC tissues and cell lines | [16] |
| LncRNA MEG3 | downregulation | anti-proliferative role in EC by repressing Notch signaling pathway | In vitro assay from EC tissues and cell lines | [38] |
| LncRNA GAS5/miR-103 | downregulation / inhibition of miRNA-103 expression | tumor suppressor | In vitro assay from EC tissues and cell lines | [40] |
| LincRNA LINC00958 | upregulation | increase of LINC00958 in EC | In vitro assay from EC tissues | [51] |
| LincRNA LINC00672 | downregulation | downregulation during EC development | In vitro assay from EC tissues | [33] |
| NcRNA Nc886 | upregulation | upregulation of nc886 in EC late phases, compared to early stages and Normal Endometrial Tissues (NET) | In vitro assay from EC tissues | [57] |
| miRNA-93 | upregulation | over-expression associated to cell migration and invasion | In vitro assay from EC tissues and cell lines | [31] |
| miRNA-205 | upregulation | prognostic marker associated with better overall survival | In vitro assay from EC tissues | [52] |
| miRNA-944 | upregulation | over-expression in EC tissues compared to NET | In vitro assay from EC tissues | [54] |
| miRNA-29b | miRNA-29b/MAPK/ERK and PI3K/Akt signaling pathways | inhibition of angiogenesis by targeting VEGFA through MAPK/ERK and PI3K/Akt signaling | In vitro assay from EC tissues and cell lines | [62] |
| miRNA-145 | upregulation | upregulation of miR-145 lead to downregulation of linc-RoR7Dicer | In vitro assay from EC tissues | [49] |
| miRNA-361 | miR-361/twist signaling and miR-361/let-7b downregulation | lower let-7b, twist signaling and miR-361 are associated with worse patient outcome | In vitro assay from EC tissues | [60] |
| miRNA-194 | downregulation | downregulation associated to EC poor prognosis | In vitro assay from EC tissues and cell lines | [62] |
References
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488.
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933.
- Ling, H.; Vincent, K.; Pichler, M.; Fodde, R.; Berindan-Neagoe, I.; Slack, F.J.; Calin, G.A. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene 2015, 34, 5003–5011.
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007, 17, 556–565.
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914.
- Smolle, M.A.; Bullock, M.D.; Ling, H.; Pichler, M.; Haybaeck, J. Long Non-Coding RNAs in Endometrial Carcinoma. Int. J. Mol. Sci. 2015, 16, 26463–26472.
- Mattick, J.S.; Amaral, P.P.; Dinger, M.E.; Mercer, T.R.; Mehler, M.F. RNA regulation of epigenetic processes. Bioessays 2009, 31, 51–59.
- Cheetham, S.W.; Gruhl, F.; Mattick, J.S.; Dinger, M.E. Long noncoding RNAs and the genetics of cancer. Br. J. Cancer 2013, 108, 2419–2425.
- Lv, Z.; Xu, Q.; Yuan, Y. A systematic review and meta-analysis of the association between long non-coding RNA polymorphisms and cancer risk. Rev. Mutat. Res. 2017, 771, 1–14.
- Pereira, D.M.; Rodrigues, P.M.; Borralho, P.M.; Rodrigues, C.M. Delivering the promise of miRNA cancer therapeutics. Drug Discov. Today 2013, 18, 282–289.
- McCaskill, J.; Praihirunkit, P.; Sharp, P.M.; Buck, A.H. RNA-mediated degradation of microRNAs: A widespread viral strategy? RNA Biol. 2015, 12, 579–585.
- Sheng, X.; Li, J.; Yang, L.; Chen, Z.; Zhao, Q.; Tan, L.; Zhou, Y.; Li, J. Promoter hypermethylation influences the suppressive role of maternally expressed 3, a long non-coding RNA, in the development of epithelial ovarian cancer. Oncol. Rep. 2014, 32, 277–285.
- Sun, K.X.; Wu, D.D.; Chen, S.; Zhao, Y.; Zong, Z.H. LncRNA MEG3 inhibit endometrial carcinoma tumorigenesis and progression through PI3K pathway. Apoptosis 2017, 22, 1543–1552.
- Li, Q.; Zhang, C.; Chen, R.; Xiong, H.; Qiu, F.; Liu, S.; Zhang, M.; Wang, F.; Wang, Y.; Zhou, X.; et al. Disrupting MALAT1/miR-200c sponge decreases invasion and migration in endometrioid endometrial carcinoma. Cancer Lett. 2016, 383, 28–40.
- Vennin, C.; Spruyt, N.; Dahmani, F.; Julien, S.; Bertucci, F.; Finetti, P.; Chassat, T.; Bourette, R.P.; Le Bourhis, X.; Adriaenssens, E. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget 2015, 6, 29209–29223.
- Li, Z.; Wei, D.; Yang, C.; Sun, H.; Lu, T.; Kuang, D. Overexpression of long noncoding RNA NEAT1 promotes cell proliferation, invasion and migration in endometrial endometrioid adenocarcinoma. Biomed. Pharmacother. 2016, 84, 244–251.
- Palmieri, G.; Paliogiannis, P.; Sini, M.C.; Manca, A.; Palomba, G.; Doneddu, V.; Tanda, F.; Pascale, M.R.; Cossu, A. Long non-coding RNA CASC2 in human cancer. Crit. Rev. Oncol. Hematol. 2017, 111, 31–38.
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927.
- Sun, M.Y.; Zhu, J.Y.; Zhang, C.Y.; Zhang, M.; Song, Y.N.; Rahman, K.; Zhang, L.J.; Zhang, H. Autophagy regulated by lncRNA HOTAIR contributes to the cisplatin-induced resistance in endometrial cancer cells. Biotechnol. Lett. 2017, 39, 1477–1484.
- Akhmedkhanov, A.; Zeleniuch-Jacquotte, A.; Toniolo, P. Role of exogenous and endogenous hormones in endometrial cancer: Review of the evidence and research perspectives. Ann. N. Y. Acad. Sci. 2001, 943, 296–315.
- Bhan, A.; Hussain, I.; Ansari, K.I.; Kasiri, S.; Bashyal, A.; Mandal, S.S. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J. Mol. Biol. 2013, 425, 3707–3722.
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076.
- He, X.; Bao, W.; Li, X.; Chen, Z.; Che, Q.; Wang, H.; Wan, X.P. The long non-coding RNA HOTAIR is upregulated in endometrial carcinoma and correlates with poor prognosis. Int. J. Mol. Med. 2014, 33, 325–332.
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schütz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P.; et al. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835–839.
- Lanz, R.B.; Chua, S.S.; Barron, N.; Söder, B.M.; DeMayo, F.; O’Malley, B.W. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol. Cell. Biol. 2003, 23, 7163–7176.
- Li, L.; Gu, M.; Bo, Y.; Shi, S.; Shan, Y.; Bao, L.; Yiwen, Y. Long non-coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinoma. Cancer Sci. 2016, 107, 1215–1222.
- Xu, J.; Qian, Y.; Ye, M.; Fu, Z.; Jia, X.; Li, W.; Xu, P.; Lv, M.; Huang, L.; Wang, L.; et al. Distinct expression profile of lncRNA in endometrial carcinoma. Oncol. Rep. 2016, 36, 3405–3412.
- Jiang, Y.; Malouf, G.G.; Zhang, J.; Zheng, X.; Chen, Y.; Thompson, E.J.; Weinstein, J.N.; Yuan, Y.; Spano, J.P.; Broaddus, R.; et al. Long non-coding RNA profiling links subgroup classification of endometrioid endometrial carcinomas with trithorax and polycomb complex aberrations. Oncotarget 2015, 6, 39865–39876.
- Liu, L.; Chen, X.; Zhang, Y.; Hu, Y.; Shen, X.; Zhu, W. Long non-coding RNA TUG1 promotes endometrial cancer development via inhibiting miR-299 and miR-34a-5p. Oncotarget 2017, 9, 31386–31394.
- Yang, L.; Zhang, J.; Jiang, A.; Liu, Q.; Li, C.; Yang, C.; Xiu, J. Expression profile of long non-coding RNAs is altered in endometrial cancer. Int. J. Clin. Exp. Med. 2015, 8, 5010–5021.
- Chen, S.; Chen, X.; Sun, K.X.; Xiu, Y.L.; Liu, B.L.; Feng, M.X.; Sang, X.B.; Zhao, Y. MicroRNA-93 promotes epithelial-mesenchymal transition of endometrial carcinoma cells. PLoS ONE 2016, 11, e0165776.
- Zhai, W.; Li, X.; Wu, S.; Zhang, Y.; Pang, H.; Chen, W. Microarray expression profile of lncRNAs and the upregulated ASLNC04080 lncRNA in human endometrial carcinoma. Int. J. Oncol. 2015, 46, 2125–2137.
- Li, W.; Li, H.; Zhang, L.; Hu, M.; Li, F.; Deng, J.; An, M.; Wu, S.; Ma, R.; Lu, J.; et al. Long non-coding RNA LINC00672 contributes to p53 protein-mediated gene suppression and promotes endometrial cancer chemosensitivity. J. Biol. Chem. 2017, 292, 5801–5813.
- Sun, Y.; Zou, X.; He, J.; Mao, Y. Identification of long non-coding RNAs biomarkers associated with progression of endometrial carcinoma and patient outcomes. Oncotarget 2017, 8, 52604–52613.
- Zhao, L.; Li, Z.; Chen, W.; Zhai, W.; Pan, J.; Pang, H.; Li, X. H19 promotes endometrial cancer progression by modulating epithelial-mesenchymal transition. Oncol. Lett. 2017, 13, 363–369.
- Kaufhold, S.; Bonavida, B. Central role of Snail1 in the regulation of EMT and resistance in cancer: A target for therapeutic intervention. J. Exp. Clin. Cancer Res. 2014, 33, 62.
- Wang, D.; Wang, D.; Wang, N.; Long, Z.; Ren, X. Long Non-Coding RNA BANCR promotes endometrial cancer cell proliferation and invasion by regulating MMP2 and MMP1 via ERK/MAPK signaling pathway. Cell. Physiol. Biochem. 2016, 40, 644–656.
- Guo, Q.; Qian, Z.; Yan, D.; Li, L.; Huang, L. LncRNA-MEG3 inhibits cell proliferation of endometrial carcinoma by repressing Notch signaling. Biomed. Pharmacother. 2016, 82, 589–594.
- Shang, C.; Lang, B.; Ao, C.N.; Meng, L. Long non-coding RNA tumor suppressor candidate 7 advances chemotherapy sensitivity of endometrial carcinoma through targeted silencing of miR-23b. Tumour Biol. 2017, 39.
- Guo, C.; Song, W.Q.; Sun, P.; Jin, L.; Dai, H.Y. LncRNA-GAS5 induces PTEN expression through inhibiting miR-103 in endometrial cancer cells. J. Biomed. Sci. 2015, 22, 100.
- Qiao, Q.; Li, H. LncRNA FER1L4 suppresses cancer cell proliferation and cycle by regulating PTEN expression in endometrial carcinoma. Biochem. Biophys. Res. Commun. 2016, 478, 507–512.
- Feng, S.; Yao, J.; Chen, Y.; Geng, P.; Zhang, H.; Ma, X.; Zhao, J.; Yu, X. Expression and functional role of reprogramming-related long noncoding RNA (lincRNA-ROR) in Glioma. J. Mol. Neurosci. 2015, 56, 623–630.
- Eades, G.; Wolfson, B.; Zhang, Y.; Li, Q.; Yao, Y.; Zhou, Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol. Cancer Res. 2015, 13, 330–338.
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014, 5, e1287.
- Gao, S.; Wang, P.; Hua, Y.; Xi, H.; Meng, Z.; Liu, T.; Chen, Z.; Liu, L.M. ROR functions as a ceRNA to regulate Nanog expression by sponging miR-145 and predicts poor prognosis in pancreatic cancer. Oncotarget 2016, 7, 1608–1618.
- Zhan, H.X.; Wang, Y.; Li, C.; Xu, J.W.; Zhou, B.; Zhu, J.K.; Han, H.F.; Wang, L.; Wang, Y.S.; Hu, S.Y. LincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer Lett. 2016, 374, 261–271.
- Takahashi, K.; Yan, I.K.; Haga, H.; Patel, T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J. Cell Sci. 2014, 127, 1585–1594.
- Takahashi, K.; Yan, I.K.; Kogure, T.; Haga, H.; Patel, T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio 2014, 4, 458–467.
- Zhou, X.; Gao, Q.; Wang, J.; Zhang, X.; Liu, K.; Duan, Z. Linc-RNA-RoR acts as a “sponge” against mediation of the differentiation of endometrial cancer stem cells by microRNA-145. Gynecol. Oncol. 2014, 133, 333–339.
- Rezaei, M.; Emadi-Baygi, M.; Hoffmann, M.J.; Schulz, W.A.; Nikpour, P. Altered expression of LINC-ROR in cancer cell lines and tissues. Tumour Biol. 2016, 37, 1763–1769.
- Chen, B.J.; Byrne, F.L.; Takenaka, K.; Modesitt, S.C.; Olzomer, E.M.; Mills, J.D.; Farrell, R.; Hoehn, K.L.; Janitz, M. Transcriptome landscape of long intergenic non-coding RNAs in endometrial cancer. Gynecol. Oncol. 2017, 147, 654–662.
- Wilczynski, M.; Danielska, J.; Dzieniecka, M.; Szymanska, B.; Wojciechowski, M.; Malinowski, A. Prognostic and clinical significance of miRNA-205 in endometrioid endometrial cancer. PLoS ONE 2016, 11, e0164687.
- Jayaraman, M.; Radhakrishnan, R.; Mathews, C.A.; Yan, M.; Husain, S.; Moxley, K.M.; Song, Y.S.; Dhanasekaran, D.N. Identification of novel diagnostic and prognostic miRNA signatures in endometrial cancer. Genes Cancer 2017, 8, 566–576.
- He, Z.; Xu, H.; Meng, Y.; Kuang, Y. miR-944 acts as a prognostic marker and promotes the tumor progression in endometrial cancer. Biomed. Pharmacother. 2017, 88, 902–910.
- Ravo, M.; Cordella, A.; Rinaldi, A.; Bruno, G.; Alexandrova, E.; Saggese, P.; Nassa, G.; Giurato, G.; Tarallo, R.; Marchese, G.; et al. Small non-coding RNA deregulation in endometrial carcinogenesis. Oncotarget 2015, 6, 4677–4691.
- Ahsen, M.E.; Boren, T.P.; Singh, N.K.; Misganaw, B.; Mutch, D.G.; Moore, K.N.; Backes, F.J.; McCourt, C.K.; Lea, J.S.; Miller, D.S.; et al. Sparse feature selection for classification and prediction of metastasis in endometrial cancer. BMC Genom. 2017, 27, 233.
- Bahubeshi, A.; Tischkowitz, M.; Foulkes, W.D. miRNA processing and human cancer: DICER1 cuts the mustard. Sci. Transl. Med. 2011, 3.
- Hu, Z.; Zhang, H.; Tang, L.; Lou, M.; Geng, Y. Silencing nc886, a non-coding RNA, induces apoptosis of human endometrial cancer cells-1A in vitro. Med. Sci. Monit. 2017, 16, 1317–1324.
- The Cancer Genome Atlas; National Cancer Institute, National Human Genome Research Institute. Available online: (accessed on 27 March 2018).
- Ihira, K.; Dong, P.; Xiong, Y.; Watari, H.; Konno, Y.; Hanley, S.J.; Noguchi, M.; Hirata, N.; Suizu, F.; Yamada, T.; et al. EZH2 inhibition suppresses endometrial cancer progression via miR-361/Twist axis. Oncotarget 2017, 8, 13509–13520.
- Gong, B.; Yue, Y.; Wang, R.; Zhang, Y.; Jin, Q.; Zhou, X. Overexpression of microRNA-194 suppresses the epithelial-mesenchymal transition in targeting stem cell transcription factor Sox3 in endometrial carcinoma stem cells. Tumor Biol. 2017, 39.
- Chen, H.X.; Xu, X.X.; Tan, B.Z.; Zhang, Z.; Zhou, X.D. MicroRNA-29b inhibits angiogenesis by targeting VEGFA through the MAPK/ERK and PI3K/Akt signaling pathways in endometrial carcinoma. Cell. Physiol. Biochem. 2017, 41, 933–946.
- Alowayed, N.; Salker, M.S.; Zeng, N.; Singh, Y.; Lang, F. LEFTY2 controls migration of human endometrial cancer cells via focal adhesion kinase activity (FAK) and miRNA-200a. Cell. Physiol. Biochem. 2016, 39, 815–826.
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36.
- Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 2008, 358, 1148–1159.
- Hosseini, E.S.; Meryet-Figuiere, M.; Sabzalipoor, H.; Kashani, H.H.; Nikzad, H.; Asemi, Z. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol. Cancer 2017, 16, 107.
- Ivanga, M.; Labrie, Y.; Calvo, E.; Belleau, P.; Martel, C.; Luu-The, V.; Morissette, J.; Labrie, F.; Durocher, F. Temporal analysis of E2 transcriptional induction of PTP and MKP and downregulation of IGF-I pathway key components in the mouse uterus. Physiol. Genom. 2007, 29, 13–23.
