Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Genetics & Heredity
Poly ADP-ribose polymerases (PARP) are specialized enzymes that catalyze the addition of ADP ribose units from “nicotinamide adenine dinucleotide-donor molecules” to their target substrates.
- PARP1
- ADP-ribosyl transferases
- PARylation
1. Introduction
ADP-ribosylation (ADPr) is the most frequent protein post-translational modification (PTM) in eukaryotes. ADPr is mediated by ADP-ribosyl transferases (ADPRT) that covalently attach single or multiple ADP-ribose units from a “donor” molecule the nicotinamide adenine dinucleotide (NAD) onto an “acceptor” substrate (i.e., proteins, nucleic acids or other small molecules). Poly ADP-ribose polymerases (PARPs) are ADPRT producing chains of ADP-ribose polymers (PAR) of variable sizes (from 2 to more than 200 units) and structures (linear and branched) [1][2] through a process known as PARylation. PARylation is involved in a plethora of biological processes that control the cell fate including: chromatin remodeling, DNA methylation changes and gene expression [3][4][5].
2. The PARP Family Members
The PARP family consists of seventeen enzymes categorized into four subfamilies and classified according to their structures and domains a representative scheme is shown in Figure 1.
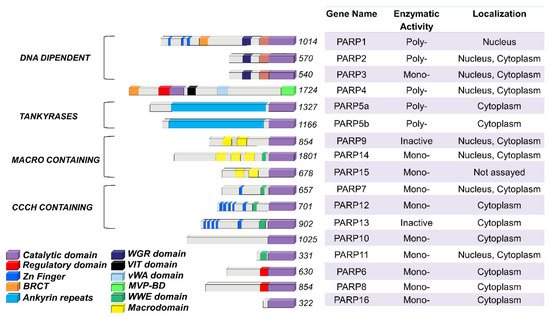
Figure 1. PARP family members functional domains. The structures are schematized in colored bars and their specific enzymatic activity including: Mono-, Poly-(ADP)ribose polymerases or Inactive is also reported.
The DNA-dependent PARPs include PARP1, PARP2, and PARP3. These are activated by discontinuous DNA structures through their amino-terminal DNA-binding domains [6]. The tankyrase subfamily, comprised of tankyrase 1 (also known as PARP5A) and tankyrase 2 (also known as PARP5B), is characterized by ankyrin domain repeats. The ankyrin-domain is a 33-residue motif consisting of two α helices separated by loops. It is responsible for protein protein interactions [7]. The CCCH subfamily contains Cys-Cys-Cys-His zinc-finger motifs that bind the RNA (PARP7, PARP12 and PARP13) and a WWE sequence, consisting of Trp-Trp-Glu domains, possesses PAR-binding activity (PARP14, PARP7, PARP12, PARP13, PARP11). The fourth and final subfamily, macro PARPs, possess ADP-ribose-binding macro domain (PARP9, PARP14, PARP15).
Of the PARP members, PARP1 has been extensively studied and is translated to clinical use. Identified over 50 years ago, PARP1 is the most abundant PARP isoform, localized predominantly in the nucleus [8][9]. The human PARP1 is a 113 KDa protein with a modular structure composed of multiple independent domains: the DNA-binding domain (DBD; residues 1–374), the auto-modification domain (residues 375–525), and the catalytic domain (residues 526–1014). The DBD at the N-terminal end contains two zinc finger motifs (Zn1 and Zn2) that are able to bind to DNA structures [10], nuclear localization signals [11], and a caspase-3 cleavage site [12]. The auto-modification domain includes the typical fold recurrent in DNA repair proteins—a BRCA1 C-terminus (BRCT) arrangement that mediates protein protein interactions and recruits DNA repair enzymes [3]. The catalytic domain of PARP1 at the C-terminal of the primary protein structure is highly conserved in the PARP superfamily, particularly within the NAD binding region [13].
3. Basic Notions of PARylation
PARPs catalyze the transfer of ADP-ribose units from NAD+ to specific target proteins, modulating their biological functions. This process, known as PARylation, generates one ADP-ribose and one nicotinamide per molecule of NAD converted. The ADP-ribose unit is then attached on carboxyl group from Glu, Asp and/or Lys residues in the target protein structure [14]. The bond formation between ADP-ribose units during PARylation occurs by either elongation or branching a schematic of the reaction is shown in Figure 2.
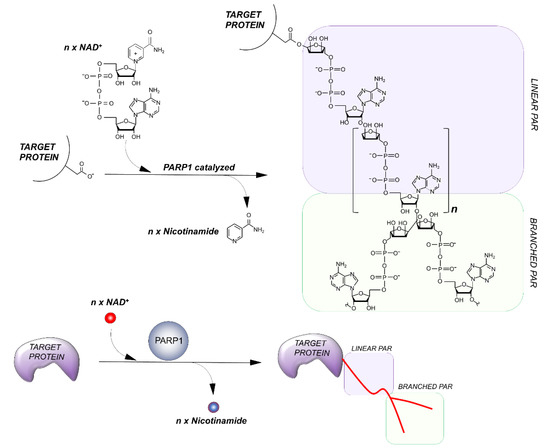
Figure 2. Outline of the PARylation reaction. The elongation reaction results in the production of a linear PAR chain; branching instead induces the rising of collateral branches.
During elongation, adenine-proximal ribose units attach to the α (1 → 2) O-glycosidic bond and produce a linear PAR chain. During branching, nicotinamide-proximal ribose units induce the rising of collateral branches. Elongation reactions generate PAR-polymers composed of more than 200 units. Branching reactions occur with less frequency, about once for every 20 elongation reactions [15].
Not all PARP members are capable of carrying out PARylation. Those acting as mono ADP-ribosyl transferases are generally referred as ADP-ribosyl-transferases (ARTDs). Other PARP members lack enzymatic activity entirely. While PARP1, PARP2, vPARP (vault PARP; also known as PARP4), PARP5A, and PARP5B catalyze PARylation, PARP3, PARP10, PARP14 and PARP15 are mono ADP-ribosyl transferases. PARP16 and 17 are catalytically inactive, mediating ADP-ribosylation through interactions with specific yet often unknown cofactors [16][17]. Although features of PARylation have been well defined for PARP1 and PARP2, the variety of mechanisms of other PARP members remains poorly understood.
4. Mechanisms and Clinical Applications of PARP Inhibitors
PARP enzymes have been shown to play a significant role in DDR by recruiting and PARylating various enzymes. PARP inhibitors are nicotinamide analogs that function by competitively binding to the NAD+ binding site on both PARP1 and PARP2 [18] A list of PARP inhibitors is reported in Table 1. Due to PARP’s role in DDR, these inhibitors can find clinical application to increase cytotoxicity in malignant cells by decreasing their ability to repair damaged DNA [19]. PARP inhibitors have been particularly successful in treating germline BRCA mutated cancers [18]. As of January 2021, there are four Food and Drug Administration (FDA) approved PARP inhibitors recommended for the treatment of various cancers (Table 1). A fifth drug, veliparib, is currently in Phase III clinical trials and is showing promising results [18].
Table 1. PARP inhibitors and their clinical uses.
| Name | Description | Reference |
|---|---|---|
| Olaparib | In HER-2 negative metastatic breast cancer patients with a germline BRCA mutation, olaparib has been shown to be very effective. Response rate of 59.9% compared to 28.8% in the standard therapy group. | [20]. |
| Niraparib | In patients with platinum sensitive recurrent ovarian cancer, niraparib greatly enhanced progression-free survival as compared to placebo. These results were consistent regardless of a germline BRCA mutation or homologous recombination deficiency (HRD) status. | [21] |
| Rucaparib | Rucaparib is generally a third (or later) line treatment used in patients with BRCA mutated ovarian cancer and as maintenance therapy for patients with recurrent or relapsed platinum sensitive ovarian cancer. Analysis has revealed an objective response rate of 54%. | [22] |
| Talazoparib | Used in patients with advanced breast cancer and germline BRCA mutations. Talazoparib has shown a significantly higher likelihood of progression-free survival (62.6% compared to 27.2% in the standard therapy group). | [23] |
PARP inhibitors are most often used in combination with other targeted therapies and chemotherapy. Rucaparib, for example, is commonly used as a maintenance therapeutic alongside platinum-based chemotherapy (cite 30830551). A January 2021 study found that olaparib treatment coupled with stimulator of interferon genes (STING) agonism induces more significant STING activation than STING agonism alone [24]. By increasing cytotoxic T cell response, this combined therapy improved anti-tumor effects significantly [24]. Furthermore, this effect is more significant when also coupled with checkpoint inhibitors that block PD-1 [25].
Similarly, olaparib is used in accordance with chemotherapy and radiation. Interestingly, olaparib’s cytotoxic effects potentiate the effects of chemo and radiation therapy [26]. PARP inhibition is far more cytotoxic than simple knockout of PARP genes [27]. This increase in cytotoxicity can be explained via a process known as “trapping”. Through an unknown mechanism, PARP inhibitors lock PARP1 and PARP2 at the site of the damaged DNA [27]. These trapped enzymes further prevent other DDR enzymes from translocating to the damaged DNA, further increasing cytotoxicity. Alongside the decrease in catalytic activity, PARP inhibitors exhibit a two-sided attack on malignant cells.
These effects also make the targeted cells more susceptible to chemotherapy and radiation therapy. Alongside the decrease in catalytic activity, PARP inhibitors exhibit a maintenance of heterochromatin throughout cell divisions by controlling UHRF1-DNMT1 interplay and by directing DNMT1 to euchromatin regions and hemi-methylated CpG dyads [22]. The interaction of UHRF1-PARP1 seems also essential for cell viability, as recent findings suggest its involvement in response to DNA damage [23].
Though extremely effective, PARP inhibitor resistance is common and arises via a multitude of mechanisms. BRCA deficient malignancies develop resistance via restoration of homologous recombination (HR) repair [28]. Resistance can also develop through stabilization of replication forks [28]. In the case of rucaparib, sensitivity to the drug in high-grade serous ovarian carcinoma (HGSOC) is determined by the methylation of the BRCA gene. Homozygous methylation predicts improved response to the drug, whereas hemizygous methylation correlates to resistance [29].
This entry is adapted from the peer-reviewed paper 10.3390/genes12030446
References
- Alemasova, E.E.; Lavrik, O.I. Poly(ADP-ribosyl)ation by PARP1: Reaction mechanism and regulatory proteins. Nucleic Acids Res. 2019, 47, 3811–3827.
- Gupte, R.; Liu, Z.; Kraus, W.L. PARPs and ADP-ribosylation: Recent advances linking molecular functions to biological outcomes. Genes Dev. 2017, 31, 101–126.
- Ray Chaudhuri, A.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621.
- Kraus, W.L.; Hottiger, M.O. PARP-1 and gene regulation: Progress and puzzles. Mol. Asp. Med. 2013, 34, 1109–1123.
- Caiafa, P.; Guastafierro, T.; Zampieri, M. Epigenetics: Poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. FASEB J. 2009, 23, 672–678.
- Ame, J.C.; Rolli, V.; Schreiber, V.; Niedergang, C.; Apiou, F.; Decker, P.; Muller, S.; Hoger, T.; Menissier-de Murcia, J.; de Murcia, G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999, 274, 17860–17868.
- Sbodio, J.I.; Chi, N.W. Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner. J. Biol. Chem. 2002, 277, 31887–31892.
- Chambon, P.; Weill, J.D.; Mandel, P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 1963, 11, 39–43.
- Sugimura, T.; Miwa, M. Poly(ADP-ribose): Historical perspective. Mol. Cell. Biochem. 1994, 138, 5–12.
- Zandarashvili, L.; Langelier, M.F.; Velagapudi, U.K.; Hancock, M.A.; Steffen, J.D.; Billur, R.; Hannan, Z.M.; Wicks, A.J.; Krastev, D.B.; Pettitt, S.J.; et al. Structural basis for allosteric PARP-1 retention on DNA breaks. Science 2020, 368.
- Schreiber, V.; Molinete, M.; Boeuf, H.; de Murcia, G.; Menissier-de Murcia, J. The human poly(ADP-ribose) polymerase nuclear localization signal is a bipartite element functionally separate from DNA binding and catalytic activity. EMBO J. 1992, 11, 3263–3269.
- Kaufmann, S.H.; Desnoyers, S.; Ottaviano, Y.; Davidson, N.E.; Poirier, G.G. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: An early marker of chemotherapy-induced apoptosis. Cancer Res. 1993, 53, 3976–3985.
- Ruf, A.; Mennissier de Murcia, J.; de Murcia, G.; Schulz, G.E. Structure of the catalytic fragment of poly(AD-ribose) polymerase from chicken. Proc. Natl. Acad. Sci. USA 1996, 93, 7481–7485.
- Zhang, Y.; Wang, J.; Ding, M.; Yu, Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods 2013, 10, 981–984.
- Kiehlbauch, C.C.; Aboul-Ela, N.; Jacobson, E.L.; Ringer, D.P.; Jacobson, M.K. High resolution fractionation and characterization of ADP-ribose polymers. Anal. Biochem. 1993, 208, 26–34.
- Loseva, O.; Jemth, A.S.; Bryant, H.E.; Schuler, H.; Lehtio, L.; Karlberg, T.; Helleday, T. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J. Biol. Chem. 2010, 285, 8054–8060.
- Kleine, H.; Poreba, E.; Lesniewicz, K.; Hassa, P.O.; Hottiger, M.O.; Litchfield, D.W.; Shilton, B.H.; Luscher, B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell 2008, 32, 57–69.
- Min, A.; Im, S.A. PARP Inhibitors as Therapeutics: Beyond Modulation of PARylation. Cancers 2020, 12, 394.
- Curtin, N.J.; Szabo, C. Poly(ADP-ribose) polymerase inhibition: Past, present and future. Nat. Rev. Drug Discov. 2020, 19, 711–736.
- Robson, M.; Im, S.-A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017, 377, 1700.
- Del Campo, J.M.; Matulonis, U.A.; Malander, S.; Provencher, D.; Mahner, S.; Follana, P.; Waters, J.; Berek, J.S.; Woie, K.; Oza, A.M.; et al. Niraparib Maintenance Therapy in Patients With Recurrent Ovarian Cancer After a Partial Response to the Last Platinum-Based Chemotherapy in the ENGOT-OV16/NOVA Trial. J. Clin. Oncol. 2019, 37, 2968–2973.
- Shirley, M. Rucaparib: A Review in Ovarian Cancer. Target. Oncol. 2019, 14, 237–246.
- Litton, J.K.; Rugo, H.S.; Ettl, J.; Hurvitz, S.A.; Goncalves, A.; Lee, K.H.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M.; et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 2018, 379, 753–763.
- Pantelidou, C.; Jadhav, H.; Kothari, A.; Liu, R.; Guerriero, J.L.; Shapiro, G.I. STING agonism enhances anti-tumor immune responses and therapeutic efficacy of PARP inhibition in BRCA-associated breast cancer. bioRxiv 2021.
- Ding, L.; Kim, H.J.; Wang, Q.; Kearns, M.; Jiang, T.; Ohlson, C.E.; Li, B.B.; Xie, S.; Liu, J.F.; Stover, E.H.; et al. PARP Inhibition Elicits STING-Dependent Antitumor Immunity in Brca1-Deficient Ovarian Cancer. Cell Rep. 2018, 25, 2972–2980 e2975.
- Bochum, S.; Berger, S.; Martens, U.M. Olaparib. Recent Results Cancer Res. 2018, 211, 217–233.
- Murai, J.; Huang, S.Y.; Das, B.B.; Renaud, A.; Zhang, Y.; Doroshow, J.H.; Ji, J.; Takeda, S.; Pommier, Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012, 72, 5588–5599.
- D’Andrea, A.D. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair 2018, 71, 172–176.
- Kondrashova, O.; Topp, M.; Nesic, K.; Lieschke, E.; Ho, G.Y.; Harrell, M.I.; Zapparoli, G.V.; Hadley, A.; Holian, R.; Boehm, E.; et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat. Commun. 2018, 9, 3970.
This entry is offline, you can click here to edit this entry!
