Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Agriculture, Dairy & Animal Science
The demand for animal protein has increased considerably worldwide, especially in China, where large numbers of livestock and poultry are produced. Antibiotics have been widely applied to promote growth and prevent diseases. However, the overuse of antibiotics in animal feed has caused serious environmental and health risks, especially the wide spread of antimicrobial resistance (AMR), which seriously affects animal and human health, food safety, ecosystems, and the sustainable future development of animal protein production.
- antibiotics
- antimicrobial resistance (AMR)
- bacteria
- China
- human and animal health
- livestock and poultry manure
1. Introduction
With the increasing demand for protein-rich food, global animal production has intensified considerably, especially in developing countries. Antibiotics are widely used in animal husbandry because of their functions in promoting growth and preventing disease. Antibiotics provide benefits, but also cause severe problems to food safety, human health, and the environment, leading to the antimicrobial resistance (AMR) problem [1]. It has been reported that by 2050, an estimated 10 million deaths per year worldwide will be attributable to AMR, and the economic burden in the health sector will reach as high as USD 100 trillion [2]. Given that AMR has become a big threat to global public health, it requires a joint effort and commitment by the international community, and across sectors, to mitigate and manage it.
It is reported that approximately 80% of all antibiotics sold in the United States are sold for use in animal agriculture to boost animal growth and protect them from infections [3]. Moreover, data for antimicrobial sales collected from 41 countries showed estimated global antimicrobial sales of 93,309 tonnes in 2017 and 104,079 tonnes in 2030, with a rise of 11.5% [4]. Hence, effective mitigation of the risks related to the extensive use of antimicrobials in animal farming and management of the AMR problem are essential, and require multi-disciplinary integrated approaches, such as the “One Health” approach. “One Health” involves a collaborative, multisectoral, and trans-disciplinary approach, operating at local, regional, national, and global levels, in order to achieve optimal health and well-being outcomes and emphasizing the interconnections between people, animals, plants, and their shared ecosystem and environment (http://www.who.int (accessed on 30 March 2021)). For instance, the recent outbreaks of swine influenza, Ebola, and the devastating ongoing Covid-19 pandemic are examples of zoonotic diseases transmitted between animals (including wildlife) and humans. Luckily, many countries have already taken action to reduce the application of antimicrobials in animal production; in particular China, the world’s largest meat consumer. Already in 2016, the Chinese government launched a national pilot program coupled with policy and regulations to decrease unnecessary antimicrobial use [4]. There is unfortunately no quick fix for the AMR problem, due to its complexity. It is clear that a ban on antibiotic feed additives will directly affect the disease resistance and normal growth cycle of livestock production, thus indirectly increasing the investment costs of livestock and poultry. Hence, there is an urgent need to tackle the AMR threat and find safe and sustainable solutions to ensure the animal protein food supply and protect human, as well as animal, health in the future. This review focuses on pollution by antibiotics and AMR in livestock and poultry manure and countermeasures against this in China, in light of China’s 14th 5-year plan, which was launched recently and recognized the increasing demand for animal protein food supply and security.
Distribution of Antibiotics and Antibiotic Resistance Genes in Animal Manure
The environmental pollution caused by antibiotics and their ecotoxicological effects has become a major global problem, especially the environmental pollution caused by the abuse of antibiotics in the livestock and poultry industry [5][6][7][8], which has become one of the current international research hotspots [9]. Studies have shown that only a small amount of the administered antibiotics participate in animal metabolism and are effectively used. Most antibiotics and their antimicrobial resistance genes induced in animals are directly discharged from the body with the urine and feces of animals [10][11][12], and the AMR genes carried by animal urine and feces are subsequently transmitted to environmental microorganisms [13]. Untreated feces containing residual antibiotics and/or AMR genes are often applied to farmland as organic fertilizer, which will exert pressure on the resistance of microorganisms in the soil environment, and induce further antibiotic resistance genes, causing serious environmental pollution, food safety challenges, and ecological toxicity [14]. AMR genes are frequently detected in livestock manure [15][16][17][18]. Among these genes, the most common types of resistance genes in livestock breeding are the tetracycline resistance gene and sulfonamide resistance genes [19][20]. Chee-Sanford et al. [21] found eight tetracycline resistance genes encoding resistance in septic tanks and groundwater samples near pig farms; these were tetracycline O (tet (O)), tetracycline Q (tet (Q)), tetracycline W (tet (W)), tetracycline M (tet (M)), tetracycline b P (tetb (P)), tetracycline S (tet (S)), tetracycline T (tet (T)), and oxytetracycline (otr (A)). This investigation showed that the unregulated discharge and treatment of livestock manure will cause serious AMR gene pollution in the surrounding water and soil environment.
China is a large agricultural country, and the livestock and poultry breeding industry has gradually changed from decentralized, to intensive, farming. The use of antibiotics is common, and the antibiotic-related pollution caused by livestock and poultry manure is prominent [22][23][24]. Animal manure has become a huge antibiotic resistance gene pool. Cheng et al. [25] confirmed the existence of the tetracycline resistance gene and sulfonamide resistance gene by studying livestock farms in eastern China; He et al. [26] measured the content of antibiotic resistance genes (ARGs) in the pig manure of three commercial pig farms in southern China, and found that 22 kinds of ARGs studied were detected in almost all samples, and the abundance was high. The detection rate of sulfonamide (sul), chloramphenicol (cml), and macrolidelincosamides–streptogramin B (MLSB) resistance genes was 100%. The antibiotic residues in animal feces found in recent years are summarized in Table 1. According to the data, tetracyclines (TCS), sulfonamides (SAS), and quinolones (QNS) were commonly residual in livestock manure, and the concentration of tetracycline antibiotics was high.
Table 1. Residues of antibiotics in animal manure.
| Fecal Type | Region | Antibiotic Residues (μg/kg) | ||
|---|---|---|---|---|
| Tetracyclines | Sulfonamides | Quinolones | ||
| Pig | Tianjin | 0.08–183.5 | 0.1–32.5 | 0.1–24.7 |
| Shandong | 0.15–59.1 | 0.04–4.1 | 0.08–44.15 | |
| Shanghai | 12.27–18.70 | 4.88–7.56 | ND | |
| Beijing | 3.33–12.3 | 0.17–1.06 | 0.41–1.45 | |
| Jiangsu | 0.51–0.56 | 0.01–0.43 | ND | |
| Sichuan | 0.015–215.3 | 0.002–6.79 | 0.012–0.125 | |
| Three northeastern provinces | 0.32–56.81 | 0.1–4.84 | 0.14–3.18 | |
| Cattle | Shandong | 0.24–59.06 | 0.06–0.36 | 0.41–46.7 |
| Shanghai | 12.01–21.36 | 4.57–9.36 | ND | |
| Jiangsu | 0.52 | ND | ND | |
| Sichuan | 0.015–2.5 | 0.002–0.07 | 0.01–0.74 | |
| Three northeastern provinces | 0.21–10.37 | 0.08–1.02 | 0.61–4.17 | |
| Chicken | Shandong | 0.14–17.68 | 0.02–6.04 | ND |
| Tianjin | 0.6–173.2 | 0.3–26.4 | 0.3–21.9 | |
| Jiangsu | 8.9–65.7 | 0.75–2.18 | 8.73 | |
| Sichuan | 0.014–416.8 | 0.002–2.1 | 0.01–8.58 | |
| Three northeastern provinces | 0.54–13.39 | 0.09–7.11 | 0.13–15.43 | |
2. Distribution of Antibiotics and Antibiotic Resistance Genes in Water, Soil, and Plants
It is estimated that 30–90% of antibiotics are excreted from the body, in the form of parent compounds or main metabolites, along with animal feces or urine [27]. High concentrations of antibiotics in livestock manure can enter the soil and water environment in various ways (Figure 1), thus causing pollution to the ecological environment. Residual antibiotics can enter the soil through animal manure and urine fertilization, and accumulate in the soil, affecting soil fertility, crop chlorophyll synthesis, enzyme secretion, and root growth. Antibiotic residues also affect soil microbial community structure and activity, and induce the generation and spread of antibiotic resistant microorganisms and resistance genes. The adsorption and degradation abilities of soil with regard to different kinds of antibiotics are different, with relative rates in the order tetracyclines > fluoroquinolones > macrolides > sulfonamides. After the manure applied to farmland is absorbed by soil, some antibiotics will enter into groundwater through infiltration or runoff with soil leaching, resulting in water pollution. In addition, the residual antibiotics in livestock and poultry feces will pollute the surface water with the wastewater discharged from livestock farms or with rainwater.
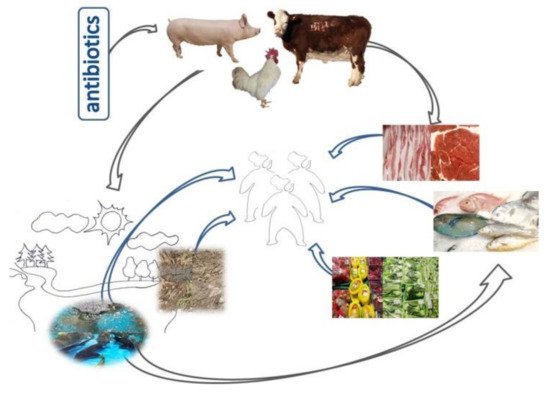
Figure 1. Veterinary antibiotics are widely used in the livestock and poultry production industry to accelerate the growth of livestock and poultry and treat diseases. This has led to the extensive spread of AMR and ARGs from animals to people, other animals, plants, the ecosystem, and the environment (soil and water pollution), as illustrated in this schematic figure.
2.1. Distribution of Antibiotics and Antibiotic Resistance Genes (ARGs) in Water
Treated or untreated aquaculture wastewater is an important source of river pollution by antibiotics and ARGs. Xu et al. [28] monitored the occurrence of nine commonly used veterinary antibiotics in the Pearl River in flood and low water seasons, including antibiotics such as ofloxacin, norfloxacin, and amoxicillin. Only amoxicillin was not detected, while the other eight kinds of antibiotics were detected, with concentration ranges of 11–67 ng/L in the flood season and 66–460 ng/L in the low water season. Nine antibiotics, except doxycycline, were detected in the sewage from livestock farms in the north, middle, and south of Jiangsu Province and in the river water near the farms, with the highest concentration ranges of 0.44–169 μg/L and 0.46–4.66 μg/L [29]. In the Yellow River and its tributaries, the average concentrations of ofloxacin, norfloxacin, roxithromycin, erythromycin, and sulfamethoxazole were 25–152 ng/L, and 44–240 ng/L in some tributaries [30]. Li et al. [29] detected seventeen kinds of antibiotics, such as ofloxacin and enrofloxacin in Baiyangdian Lake water. In addition, antibiotics have been detected in groundwater in greenhouses, where animal manure is used as organic fertilizer to grow organic vegetables.
In addition to antibiotics, a large number of ARGs were detected in the river. For example, the detection rate of sulfonamide resistance genes (sul1 and sul2) in Tianjin Haihe River was 100%, and their abundance was high [31]; there were tet A and tet B2 tetracycline resistance genes in Pearl River water, and the detection rates were 43% and 40% [32], respectively. Jiang et al. [33] detected drug resistance genes in Huangpu River and a drinking water reservoir in Shanghai, and found two sulfonamide resistance genes, nine tetracycline resistance genes, and one β—lactam resistance gene. These water bodies polluted by drug-resistant bacteria are a potential threat to human beings, and direct drinking may cause harm to human health.
The environmental risks of water pollution caused by antibiotics are now in focus and need the attention of the entire society. Antibiotics have been widely found in water, and are eventually absorbed or consumed by aquatic organisms. When antimicrobials are used in aquaculture seafood production, the AMR genes may enter into the food chain and eventually into the human body through seafood consumption. This could cause damage to human health over time and lead to drug resistance, which could have a great impact on the elderly, children, pregnant women, and other groups with weak immunity. Furthermore, because of the long-term enrichment of antimicrobials in water, some bacteria and fungi could acquire multi-drug resistance, and then form “superbugs”. Infections caused by superbugs are a true threat to human and animal health worldwide and can lead, not only to loss of life, but also to a heavy economic burden on the health sector (www.who.int (accessed on 30 March 2021)).
2.2. Distribution of Antibiotics and Antibiotic Resistance Genes in Soil
High concentrations of antibiotics can be detected in the soil around farms or after fertilizing with livestock manure. Ji et al. [34] studied the content of antibiotics in farmland soil near pig, cattle, and chicken farms in Shanghai and found the concentrations of tetracycline and oxytetracycline were 1.87–4.24 mg/kg dry matter (DM), while the concentrations of sulfadiazine, sulfamethoxazole, and sulfamethoxazole were 1.29–2.45 mg/kg DM. Hu et al. [11] investigated the content of antibiotics in the soil of organic vegetables planted with duck and pig manure in four different areas of Tianjin and found that 11 kinds of antibiotics (including tetracyclines, sulfonamides, and quinolones) were detected in the soil in winter, among which the content of oxytetracycline was the highest, reaching 2.68 mg/kg DM. Six kinds of antibiotics were detected in summer soil, among which the tetracycline content was the highest, reaching 2.5 μg/kg DM. River sediment is also often contaminated with antibiotics. Zhou et al. [35] investigated the contents of 17 kinds of antibiotics in the sediment of the Yellow River, Haihe River, and Liaohe River. The results showed that norfloxacin, enrofloxacin, ciprofloxacin, and oxytetracycline were among several antibiotics with high detection rates, and their contents reached 5770 ng/g DM, 1290 ng/g DM, 653 ng/g DM, and 652 ng/g DM, respectively.
The long-term abuse of antibiotics in the livestock and poultry breeding industry induces resistant strains in the intestinal tracts of breeding animals, and the discharge of a large amount of livestock and poultry manure will directly lead to non-point source pollution of ARGs [21]. According to Jensen et al. [36], animal manure fertilization leads to selective pressure on resistant bacteria in soil, and fertilization with animal manure is the main path for drug-resistant microorganisms and ARGs to enter the soil environment. Further studies have found that ARGs can, not only spread among strains in animal intestines, but also integrate into some mobile gene elements (such as plasmids, transposons, integrons, etc.) [37][38] and can also spread between indigenous soil bacteria and other microorganisms after entering the soil environment [39]. Under selective pressure, ARGs from pathogenic microorganisms spread among various microorganisms in the environment through gene horizontal transfer [40][41]. Moreover, heavy metals in animal feces (such as copper, zinc, and chromium) can also promote the horizontal transfer of drug-resistant genes [34]. Through this horizontal gene transfer, ARGs can migrate and transform in the soil, groundwater, and other environmental media. Owing to their strong adaptability, the indigenous microorganisms that obtain ARGs reproduce easily and become a reservoir of resistance genes [42].
2.3. Distribution of Antibiotics and Antibiotic Resistance Genes in Plants
Plants can absorb antibiotics from the soil where animal manure is applied. Hu et al. [8] detected the contents of 11 antibiotics in radish, rape, celery, and coriander planted with duck manure and pig manure as organic fertilizer in four greenhouses in four different regions of Tianjin. The results showed that ten kinds of antibiotics (0.1–57 μg/kg DM) were detected in radish, eight antibiotics (0.1–187 μg/kg DM) were detected in rape, seven antibiotics (0.1–20 μg/kg DM) were detected in celery, and seven antibiotics (0.1–532 μg/kg DM) were detected in coriander. In addition to common vegetables, antibiotics can also be detected in forage, corn, wheat, and peanut fertilized with animal manure [43]. Microorganisms in the soil can also be transferred to plants. These microorganisms transferred to plants live in the intercellular spaces or the cells of various tissues and organs of healthy plants at a certain stage, or all stages, and are called plant endophytes [44]. Many microorganisms in the soil carry drug resistance genes, which will indirectly carry resistance genes after they are transferred to plant tissues. Yang et al. [45] found that the endophytic bacteria of celery, Chinese cabbage, and cucumber planted with chicken manure generally had antibiotic resistance, and the resistance rate to cefalexin was the highest. Marti et al. [46] detected the resistance genes carried by endophytic bacteria of tomato, cucumber, pepper, radish, carrot, and lettuce planted with cow manure and pig manure. Compared with the control group, more types of ARG were detected in the group treated with animal manure. More importantly, through vegetables that can be eaten raw, ARGs in edible products and resistant plasmids in soil are likely to enter the human body along with food, thus increasing the antibiotic resistance of the human body [47]. Due to the special and hereditary nature of gene pollution, it is difficult to control and eliminate it. Once spread, it will cause long-term and irreversible harm to human health and ecosystems [48].
3. The Consequences of Antibiotic Abuse
3.1. Weaker Immune System in Livestock and Poultry
After a large number of antibiotics are ingested into the body, they will be distributed to the lymph nodes, kidney, liver, spleen, thymus, lung, bone, and other tissues and organs with blood circulation [49][50]. The immune capacity of animals will be gradually weakened, and the incidence of chronic diseases will increase. Antibiotics can also lead to the decrease of antigen quality, directly affecting the immune process, and thus have adverse effects on vaccination [51].
3.2. Cause Dysbacteriosis, Disease, or Secondary Infection in Livestock and Poultry
Although antibiotics have their own antibacterial spectrum, they not only inhibit pathogenic microorganisms, but also disturb the pattern of mutual restriction among populations or communities in the microbial flora. This leads to an imbalance of the microecology, resulting in the excessive reproduction of native bacteria or passing bacteria, further resulting in double infection or endogenous infection [52]. Especially, long-term and large-scale use of antibiotics will cause an imbalance of flora in the body; when the ecological balance is destroyed the harmful bacteria that lurk in the body take advantage of the opportunity to multiply and cause endogenous infection [53]. Antibiotics can eliminate the sensitive bacteria in the body, causing a large number of vacancies on some microbial attachment points in the body, providing opportunities for external drug-resistant bacteria to enter, thus causing exogenous infection [54].
3.3. Residues in Animal Products and the Environment
Drug residue is one of the controversies regarding adding antibiotics to feed. After being absorbed into the body, antibiotics are distributed to almost all organs of the body, especially the liver. Roughly 60–85% of antibiotics are excreted in different forms, such as feces or bile. Some antibiotics with stable properties can remain stable for a long time after being excreted into the environment, resulting in drug residues in the environment. These residual drugs are slowly accumulated in the human body and plants through animal products and the environment, and finally gathered in the human body through various pathways, resulting in the production of a large number of drug-resistant strains, loss of resistance to certain diseases, or toxic effects on the body due to a large accumulation [7].
3.4. Antibiotic Resistance of Pathogens
The formation of antibiotic-resistant pathogens is a global problem. At present, there is almost no antibiotic without antibiotic resistant pathogens. In particular, the long-term use of antibiotics in animals at doses lower than the treatment dose (such as preventive doses and growth promoting doses) can accelerate the production of antibiotic-resistant bacteria. Once antibiotic-resistant bacteria are produced, they can spread among animals, which will make animals in large-scale breeding facilities become a huge reservoir of ARGs. The antibiotic resistance of antibiotic-resistant bacteria from animals can be transferred to human beings, which has a great impact on human health. According to results published in the Proceedings of the National Academy of Sciences, 149 unique ARGs have been found in Chinese commercial pig farms, and the levels of some ARGs are 192 to 28,000 times higher than those of the control samples [4].
3.5. Negative Impact of Antibiotics and ARGs on Human Health
Previous studies have shown that penicillin, streptomycin, sulfonamides, and other antibiotics cause allergies; chloramphenicol causes regenerative, dysfunctional and hemolytic anemia, thrombocytopenia, and liver injury; tetracyclines cause photosensitivity and gastrointestinal reaction; olaquindox is a gene inducer; furazolidone induces animal carcinogenesis; aminoglycoside antibiotics cause nephrotoxicity; and fluoroquinolone keto antibiotics can cause mitochondrial damage [55][56].
The harm caused by ARGs is mainly manifested in the resistance of bacteria to antibiotics, which makes these ineffective or reduces their effect. At present, infections caused by antibiotic-resistant bacteria are becoming more common around the world [57][58]. According to a survey of infectious disease experts conducted by the emerging infection network of the United States, more than 60% of the participants said that they had seen extensive antibiotic-resistant and incurable bacterial infections in the past year [59]. The World Health Organization warned in 2014 that the antibiotic resistance of bacteria had become a terrible crisis [60]. It has been reported that by 2050, the global bacterial infectious diseases caused by antibiotic resistance are expected to cause 10 million deaths per year, 1.8 million more than those due to cancer [61].
This entry is adapted from the peer-reviewed paper 10.3390/antibiotics10050539
References
- Van Boeckel, T.P.; Glennon, E.E.; Chen, D.; Gilbert, M.; Robinson, T.P.; Grenfell, B.T.; Levin, S.A.; Bonhoeffer, S.; Laxminarayan, R. Reducing antimicrobial use in food animals. Science 2017, 357, 1350–1352.
- Tang, K.; Li, Z.; Li, W.; Chen, L. China’s Silk Road and global health. Lancet 2017, 390, 2595–2601.
- Gilbert, W.; Thomas, L.F.; Coyne, L.; Rushton, J. Review: Mitigating the risks posed by intensification in livestock production: The examples of antimicrobial resistance and zoonoses. Animal 2021, 15, 100123.
- Tiseo, K.; Huber, L.; Gilbert, M.; Robinson, T.P.; Van Boeckel, T.P. Global Trends in Antimicrobial Use in Food Animals from 2017 to 2030. Antibiotics 2020, 9, 918.
- Cabello, F.C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environ. Microbiol. 2006, 8, 1137–1144.
- McManus, P.S.; Stockwell, V.O.; Sundin, G.W.; Jones, A.L. Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 2002, 40, 443–465.
- Singer, R.S.; Finch, R.; Wegener, H.C.; Bywater, R.; Walters, J.; Lipsitch, M. Antibiotic resistance—The interplay between antibiotic use in animals and human beings. Lancet Infect. Dis. 2003, 3, 47–51.
- Smith, D.L.; Harris, A.D.; Johnson, J.A.; Silbergeld, E.K.; Morris, J.G., Jr. Animal antibiotic use has an early but important impact on the emergence of antibiotic resistance in human commensal bacteria. Proc. Natl. Acad. Sci. USA 2002, 99, 6434–6439.
- Boxall, A.B.; Fogg, L.A.; Blackwell, P.A.; Kay, P.; Pemberton, E.J.; Croxford, A. Veterinary Medicines in the Environment. In Reviews of Environmental Contamination and Toxicology; Springer: New York, NY, USA, 2004; Volume 180, pp. 1–91.
- Heuer, H.; Schmitt, H.; Smalla, K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011, 14, 236–243.
- Hu, X.; Zhou, Q.; Luo, Y. Occurrence and source analysis of typical veterinary antibiotics in manure, soil, vegetables and groundwater from organic vegetable bases, northern China. Environ. Pollut. 2010, 158, 2992–2998.
- Martinez, J.L. Antibiotics and antibiotic resistance genes in natural environments. Science 2008, 321, 365–367.
- Furushita, M.; Shiba, T.; Maeda, T.; Yahata, M.; Kaneoka, A.; Takahashi, Y.; Torii, K.; Hasegawa, T.; Ohta, M. Similarity of tetracycline resistance genes isolated from fish farm bacteria to those from clinical isolates. Appl. Environ. Microbiol. 2003, 69, 5336–5342.
- Schmitt, H.; Stoob, K.; Hamscher, G.; Smit, E.; Seinen, W. Tetracyclines and tetracycline resistance in agricultural soils: Microcosm and field studies. Microb. Ecol. 2006, 51, 267–276.
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259.
- Wiener, P.; Egan, S.; Huddleston, A.S.; Wellington, E.M. Evidence for transfer of antibiotic-resistance genes in soil populations of streptomycetes. Mol. Ecol. 1998, 7, 1205–1216.
- Xi, C.; Zhang, Y.; Marrs, C.F.; Ye, W.; Simon, C.; Foxman, B.; Nriagu, J. Prevalence of antibiotic resistance in drinking water treatment and distribution systems. Appl. Environ. Microbiol. 2009, 75, 5714–5718.
- Zhang, X.X.; Zhang, T.; Fang, H.H. Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 2009, 82, 397–414.
- Tamminen, M.; Karkman, A.; Lohmus, A.; Muziasari, W.I.; Takasu, H.; Wada, S.; Suzuki, S.; Virta, M. Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ. Sci. Technol. 2011, 45, 386–391.
- Zhang, Y.; Zhang, C.; Parker, D.B.; Snow, D.D.; Zhou, Z.; Li, X. Occurrence of antimicrobials and antimicrobial resistance genes in beef cattle storage ponds and swine treatment lagoons. Sci. Total Environ. 2013, 463–464, 631–638.
- Chee-Sanford, J.C.; Aminov, R.I.; Krapac, I.J.; Garrigues-Jeanjean, N.; Mackie, R.I. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 2001, 67, 1494–1502.
- Pallecchi, L.; Bartoloni, A.; Paradisi, F.; Rossolini, G.M. Antibiotic resistance in the absence of antimicrobial use: Mechanisms and implications. Expert Rev. AntiInfect. Ther. 2008, 6, 725–732.
- Akinbowale, O.L.; Peng, H.; Barton, M.D. Diversity of tetracycline resistance genes in bacteria from aquaculture sources in Australia. J. Appl. Microbiol. 2007, 103, 2016–2025.
- Srinivasan, V.; Nam, H.M.; Sawant, A.A.; Headrick, S.I.; Nguyen, L.T.; Oliver, S.P. Distribution of tetracycline and streptomycin resistance genes and class 1 integrons in Enterobacteriaceae isolated from dairy and nondairy farm soils. Microb. Ecol. 2008, 55, 184–193.
- Cheng, W.; Chen, H.; Su, C.; Yan, S. Abundance and persistence of antibiotic resistance genes in livestock farms: A comprehensive investigation in eastern China. Environ. Int. 2013, 61, 1–7.
- He, L.Y.; Ying, G.G.; Liu, Y.S.; Su, H.C.; Chen, J.; Liu, S.S.; Zhao, J.L. Discharge of swine wastes risks water quality and food safety: Antibiotics and antibiotic resistance genes from swine sources to the receiving environments. Environ. Int. 2016, 92–93, 210–219.
- Cheng, D.; Feng, Y.; Liu, Y.; Xue, J.; Li, Z. Dynamics of oxytetracycline, sulfamerazine, and ciprofloxacin and related antibiotic resistance genes during swine manure composting. J. Environ. Manag. 2019, 230, 102–109.
- Xu, W.H.; Zhang, G.; Zou, S.C.; Li, X.D.; Liu, Y.C. Determination of selected antibiotics in the Victoria Harbour and the Pearl River, South China using high-performance liquid chromatography-electrospray ionization tandem mass spectrometry. Environ. Pollut. 2007, 145, 672–679.
- Wei, R.; Ge, F.; Huang, S.; Chen, M.; Wang, R. Occurrence of veterinary antibiotics in animal wastewater and surface water around farms in Jiangsu Province, China. Chemosphere 2011, 82, 1408–1414.
- Xu, W.; Zhang, G.; Zou, S.; Ling, Z.; Wang, G.; Yan, W. A preliminary investigation on the occurrence and distribution of antibiotics in the Yellow River and its tributaries, China. Water Environ. Res. 2009, 81, 248–254.
- Luo, Y.; Mao, D.; Rysz, M.; Zhou, Q.; Zhang, H.; Xu, L.; Alvarez, P.J.J. Trends in antibiotic resistance genes occurrence in the Haihe River, China. Environ. Sci. Technol. 2010, 44, 7220–7225.
- Tao, R.; Ying, G.G.; Su, H.C.; Zhou, H.W.; Sidhu, J.P. Detection of antibiotic resistance and tetracycline resistance genes in Enterobacteriaceae isolated from the Pearl rivers in South China. Environ. Pollut. 2010, 158, 2101–2109.
- Jiang, L.; Hu, X.; Xu, T.; Zhang, H.; Sheng, D.; Yin, D. Prevalence of antibiotic resistance genes and their relationship with antibiotics in the Huangpu River and the drinking water sources, Shanghai, China. Sci. Total Environ. 2013, 458–460, 267–272.
- Ji, X.; Shen, Q.; Liu, F.; Ma, J.; Xu, G.; Wang, Y.; Wu, M. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai; China. J. Hazard. Mater. 2012, 235–236, 178–185.
- Zhou, L.J.; Ying, G.G.; Zhao, J.L.; Yang, J.F.; Wang, L.; Yang, B.; Liu, S. Trends in the occurrence of human and veterinary antibiotics in the sediments of the Yellow River, Hai River and Liao River in northern China. Environ. Pollut. 2011, 159, 1877–1885.
- Jensen, L.B.; Baloda, S.; Boye, M.; Aarestrup, F.M. Antimicrobial resistance among Pseudomonas spp. and the Bacillus cereus group isolated from Danish agricultural soil. Environ. Int. 2001, 26, 581–587.
- Wright, G.D. The antibiotic resistome: The nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007, 5, 175–186.
- Kruse, H.; Sorum, H. Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl. Environ. Microbiol. 1994, 60, 4015–4021.
- Rysz, M.; Alvarez, P.J. Amplification and attenuation of tetracycline resistance in soil bacteria: Aquifer column experiments. Water Res. 2004, 38, 3705–3712.
- Smalla, K.; Heuer, H.; Gotz, A.; Niemeyer, D.; Krogerrecklenfort, E.; Tietze, E. Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl. Environ. Microbiol. 2000, 66, 4854–4862.
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic resistance genes as emerging contaminants: Studies in northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450.
- Salyers, A.A.; Gupta, A.; Wang, Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004, 12, 412–416.
- Zhao, F.; Yang, L.; Chen, L.; Li, S.; Sun, L. Bioaccumulation of antibiotics in crops under long-term manure application: Occurrence, biomass response and human exposure. Chemosphere 2019, 219, 882–895.
- Wang, F.H.; Qiao, M.; Chen, Z.; Su, J.Q.; Zhu, Y.G. Antibiotic resistance genes in manure-amended soil and vegetables at harvest. J. Hazard. Mater. 2015, 299, 215–221.
- Yang, Q.; Ren, S.; Niu, T.; Guo, Y.; Qi, S.; Han, X.; Liu, D.; Pan, F. Distribution of antibiotic-resistant bacteria in chicken manure and manure-fertilized vegetables. Environ. Sci. Pollut. Res. Int. 2014, 21, 1231–1241.
- Marti, R.; Scott, A.; Tien, Y.C.; Murray, R.; Sabourin, L.; Zhang, Y.; Topp, E. Impact of manure fertilization on the abundance of antibiotic-resistant bacteria and frequency of detection of antibiotic resistance genes in soil and on vegetables at harvest. Appl. Environ. Microbiol. 2013, 79, 5701–5709.
- Shea, K.M. Antibiotic resistance: What is the impact of agricultural uses of antibiotics on children’s health? Pediatrics 2003, 112, 253–258.
- Tello, A.; Austin, B.; Telfer, T.C. Selective pressure of antibiotic pollution on bacteria of importance to public health. Environ. Health Perspect. 2012, 120, 1100–1106.
- Björnsson, E.S. Drug-induced liver injury due to antibiotics. Scand. J. Gastroenterol. 2017, 52, 617–623.
- Bergeron, M.G. Tissue penetration of antibiotics. Clin. Biochem. 1986, 19, 90–100.
- Jansen, K.U.; Anderson, A.S. The role of vaccines in fighting antimicrobial resistance (AMR). Hum. Vaccines Immunother. 2018, 14, 2142–2149.
- Francino, M.P. Antibiotics and the Human Gut Microbiome: Dysbioses and Accumulation of Resistances. Front. Microbiol. 2015, 6, 1543.
- Isaac, S.; Scher, J.U.; Djukovic, A.; Jiménez, N.; Littman, D.R.; Abramson, S.B.; Pamer, E.G.; Ubeda, C. Short- and long-term effects of oral vancomycin on the human intestinal microbiota. J. Antimicrob. Chemother. 2017, 72, 128–136.
- Neuman, H.; Forsythe, P.; Uzan, A.; Avni, O.; Koren, O. Antibiotics in early life: Dysbiosis and the damage done. FEMS Microbiol. Rev. 2018, 42, 489–499.
- Dancer, S.J. How antibiotics can make us sick: The less obvious adverse effects of antimicrobial chemotherapy. Lancet Infect. Dis. 2004, 4, 611–619.
- Marchant, J. When antibiotics turn toxic. Nature 2018, 555, 431–433.
- Golkar, Z.; Bagasra, O.; Pace, D.G. Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries. 2014, 8, 129–136.
- Barlow, G. Clinical challenges in antimicrobial resistance. Nat. Microbiol. 2018, 3, 258–260.
- Spellberg, B.; Gilbert, D.N. The future of antibiotics and resistance: A tribute to a career of leadership by John Bartlett. Clin. Infect. Dis. 2014, 59, S71–S75.
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The antimicrobial resistance crisis: Causes, consequences, and management. Front. Public Health 2014, 2, 145.
- McAllister, T.A.; Wang, Y.; Diarra, M.S.; Alexander, T.; Stanford, K. Challenges of a one-health approach to the development of alternatives to antibiotics. Anim. Front. 2018, 8, 10–20.
This entry is offline, you can click here to edit this entry!
