Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Others
Ubiquitylation and deubiquitylation are reversible protein post-translational modification (PTM) processes involving the regulation of protein degradation under physiological conditions. Loss of balance in this regulatory system can lead to a wide range of diseases, such as cancer and inflammation. As the main members of the deubiquitinases (DUBs) family, ubiquitin-specific peptidases (USPs) are closely related to biological processes through a variety of molecular signaling pathways, including DNA damage repair, p53 and transforming growth factor-β (TGF-β) pathways.
- Ubiquitin-Specific Peptidases
- Signaling Pathways
- Drug Screening
- USP Inhibitors
1. Introduction
As a complex regulatory mechanism of biological functions, post-translational modification (PTM) is essential for cell growth and stress response. Generally, intracellular proteins will experience multiple types of modifications after translation, such as phosphorylation, acetylation, methylation, and ubiquitylation, each corresponding to one or more specific functions [1]. Among them, ubiquitylation is responsible for regulating protein–protein interactions, cellular localization, and enzymatic activities of its protein substrates, and it is also related to proteasome-mediated protein degradation. A large number of studies have identified the ubiquitin-driven degradation pathways as one of the most important ways to help maintain protein balance within eukaryotic cells [1,2]. Therefore, the ubiquitylation of proteins plays indispensable regulatory roles in various biological phenomena [2].
Eukaryotic cells are equipped to recognize and degrade proteins by the ubiquitin–proteasome system (UPS). Upon conjugated to chains of ubiquitin, proteins are then directed to the 26S proteasome, a macromolecular protease, and degraded [3]. Ubiquitin is a small peptide (8.5 kDa) consisting of 76 amino acids that is ubiquitous in eukaryotic cells. The peptide sequence is highly conserved and contains seven lysine sites (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63), a glycine site at the C-terminus, and a methionine at the N-terminus (Met1). In general, the poly-ubiquitin chain linked by Lys48 is a degradation marker for proteasome, while the Lys63-linked poly-ubiquitin chain usually works with non-proteasome pathways (such as DNA repair, DNA replication, and cell signal transduction) [4,5,6]. It has been reported that the ubiquitin chains connected to the target protein through Lys6, Lys11, Lys27, Lys29 or Lys33 are also related to proteasome-mediated degradation [7]. In addition, under certain circumstances Lys63-linked ubiquitin chains can also bind and target proteins that need to be degraded by the proteasome [8].
Similar to other PTMs, the ubiquitin modification of protein is a dynamic and reversible process. Ubiquitin modification can be removed by a series of ubiquitin-specific proteases, which is called deubiquitylation. These proteases are named deubiquitinases (DUBs). Deubiquitinases specifically recognize and excise the tumbling molecules on the target protein, and also participate in the editing of poly-ubiquitin, thus playing an important role in the cleavage of ubiquitin precursors and ubiquitin monomers [9,10]. DUBs also regulate gene expression, apoptosis, cell cycle, DNA repair, and cytokines [11,12,13,14,15].
There are nearly a hundred known DUBs including cysteine proteases (USPs, UCHs, MJDs and OTUs) and metalloproteinases (containing metal catalytic domains) according to different catalytic mechanisms. They are divided into the following superfamilies: ubiquitin-specific protease (USP), ubiquitin C-terminal hydrolase (UCH), ovarian tumor protease (OTU), Machado-Josephin domain superfamily (MJD), and zinc-containing metalloproteases [16,17].
In recent years, the vast majority of DUBs have been shown to be associated with a variety of diseases, including cancer, diabetes, neurodegenerative diseases, and infectious diseases [18,19,20,21]. As the largest superfamily with over 50 members, USPs have aroused increasing attention as potential therapeutic targets in recent years [22]. It is interesting to compare USPs with kinases as drug targets since they are both involved in protein posttranslational modifications. Discovery in protein phosphorylation was awarded the Nobel Prize in 1992, and in the past thirty years numerous efforts have been invested in kinase inhibitors which resulted in a good number of clinically approved drugs including Gleevec. However, the research in the ubiquitin system caught the attention of medicinal chemists much later, and only in the past ten years USPs inhibitors have started to gradually emerge. We envision that USPs represent a new reservoir of therapeutic targets, which will reach its prime time in the twenty years to come. To date, no USP inhibitor has yet been approved for clinical use. In this review, we focus on advances in the development of USP inhibitors within the past decade.
2. Roles of USPs in Cancers
Aberrant regulation of protein ubiquitylation is closely related to the occurrence and development of tumorigenesis and other pathologies such as neurodegenerative diseases, autoimmunity, inflammatory disorders, infection, muscle dystrophies, etc. [30]. Given that the target proteins for USPs contain a large number of cell homeostasis regulators, as well as products of known oncogenes or tumor suppressor genes, USPs might be attractive and promising targets for the development of novel cancer therapies.
Studies have shown the involvement of USPs in the regulation of multiple known cancer-related pathways, including p53, transforming growth factor-β (TGF-β), protein kinase B (Akt), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), Janus kinase/signal transducers and activators of transcription (JAKs/STATs), and G protein-coupled receptor (GPCR). For example, the overexpression of USP2a stabilizes p53-murine double minute 2 (MDM2) through direct deubiquitylation, without reducing MDM2-mediated p53 ubiquitylation, and thus enhances p53 degradation [31]. Since p53 functions as a tumor suppressor and is vital for normal cellular process controlling, such downregulation of p53 can ultimately cause tumor progression [31]. USP7, however, deubiquitylates both MDM2 and p53, while its affinity to MDM2 is confirmed to be higher [32,33]. Another notable example is USP26. It has been reported to be a novel negative regulator of the TGF-β pathway and the loss of USP26 expression may be an important factor in glioblastoma pathogenesis and breast cancer [34]. Low levels of USP26 degrade drosophila mothers against decapentaplegic protein 7 (SMAD7) and stabilize TGFβ, while high levels of USP26 stabilize SMAD7 by deubiquitylation and form a complex with SMAD ubiquitylation regulatory factor 2 (SMURF2), which degrades the TGF-β receptor by ubiquitylation [34].
Here, in Table 1 we summarized the roles of USPs implicated in tumorigenesis according to the different signaling pathways on which they act.
Table 1. Cancer-related pathways regulated by USPs.
| Pathway | USPs Involved | Refs |
|---|---|---|
| DNA damage repair | USP1, USP28 | [35,36] |
| TGF-β | USP2a, USP4, USP9X, USP15, USP26 | [34,37,38,39,40] |
| Wnt/β-catenin | USP4, USP5, USP9X, USP14 | [41,42,43,44,45] |
| p53 | USP2, USP4, USP5, USP7, USP10, USP15, USP24, USP42 | [31,32,33,46,47,48,49,50,51,52,53,54,55] |
| c-Myc | USP2, USP10, USP22, USP28, USP36, USP37 | [56,57,58,59,60,61] |
| Akt | USP4, USP12, USP14, USP22, USP46 | [62,63,64,65,66,67] |
| JAKs-STATs | USP7 | [68] |
| NF-κB | USP4, USP11, USP14, USP15, USP18, USP19, USP20, USP35, USP24, USP48 | [69,70,71,72,73,74,75,76,77,78,79,80,81,82] |
| GPCR | USP4, USP8, USP14, USP20, USP30 | [83,84,85,86] |
3. Methods in Screening and Identification of Inhibitors for USPs
In order to enable the continuous discovery and development of inhibitors for USPs, a number of biological testing methods have been developed to screen and identify small molecule inhibitors (Figure 3).
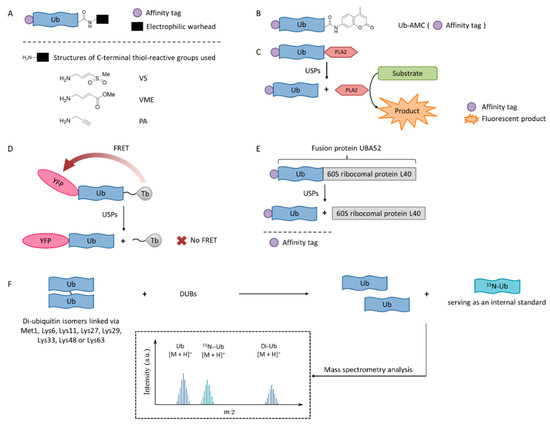
Figure 3. USPs inhibitor screening methods. (A) Activity-based probe (ABP) structures; (B) Ub-7-amino-4-methylcoumarin (AMC) structure; (C) Ub-phospholipase A2 (PLA2) assay; (D) Time-resolved fluorescence resonance energy transfer (TR-FRET) assay; (E) UBA52 structure and the SDS-PAGE-Coomassie assay; (F) Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) method.
3.1. Activity-Based Probes
Activity-based probes (ABPs), in which an electrophilic warhead is introduced onto the C-terminal glycine of ubiquitin, provide a way to test compounds in a cellular environment. Currently reported probe molecules include ubiquitin-vinylmethyl sulfone (Ub-VS) [87], ubiquitin-vinylmethyl ester (Ub-VME) [88], and ubiquitin-propargylic acid (Ub-PA) [89]. They can covalently label the nucleophilic cysteine of DUBs, resulting in a band shift on SDS-PAGE.
The advantage of this method is that it is closely related to the physiological environment of cells. The disadvantage is that it is time-consuming and laborious, so it is not currently recommended for screening USP inhibitors.
Interestingly, in order to overcome the lack of target selectivity, a novel Ub-based activity probe (Rh-M20-PA) bearing specific mutations to achieve selectivity for USP16 was developed by combining structural modelling and computation. A number of USP16-specific inhibitors were successfully discovered using these USP16-selective ABPs [90].
3.2. Ub-AMC
Ub-AMC, which has the C-terminus of a ubiquitin molecule linked to 7-amino-4-methylcoumarin (AMC), is a rather simple method, and has been widely applied in the determination of deubiquitinating enzyme activities [91]. The advantages of the Ub-AMC method are low cost per test and commercial availability. However, Ub-AMC is an unnatural substrate, and light-emitting substances (auto-fluorescence or fluorescence quenchers) can interfere with the reading. Later, researchers improved the method by replacing AMC with rhodamine-110 (Rho110) or tetramethylrhodamine, so the wavelength was red-shifted and the interference was reduced [92,93].
3.3. Ub-PLA2
In the Ub-phospholipase A2 (PLA2) method, the PLA2 does not directly emit fluorescence after being cleaved from the ubiquitin chain, but it acts on a fluorescent substrate and causes it to emit fluorescence [94]. It is also called the Ub-CHOP method and makes the screening at lower enzyme concentrations possible by amplifying the activity of deubiquitinating enzymes [95]. Its signal intensity and duration are better than the Ub-AMC. Besides, the excitation wavelength is not in the ultraviolet region, and currently there are commercial kits. However, the Ub-PLA2 method is not sensitive to some deubiquitinating enzymes of the UCH family, and its price is higher than the Ub-AMC method. USP inhibitors identified by this method include shionone and P22077 [95].
3.4. TR-FRET
The time-resolved fluorescence resonance energy transfer (TR-FRET) method is based on a full-length ubiquitin substrate that is site-specifically labeled with a yellow fluorescent protein (YFP) at the N-terminus and a terbium donor at the C-terminus. This substrate has strong fluorescence resonance energy transfer (FRET) between the two groups, while the cleavage by USPs will decrease the extent of FRET [96,97,98].
The advantage of the TR-FRET method is that it is equally sensitive to the four deubiquitinating enzymes of the UCH family. However, there is no commercial kit available.
The expansion of this application is to use diubiquitin molecules (diUb) as substrates, and this diubiquitin molecule can be connected through different lysine sites to simulate different ubiquitin chain forms [99].
3.5. SDS-PAGE-Coomassie
The development of a highly reliable assay based on a readily available SDS-PAGE-Coomassie system using UBA52 as the substrate protein has been reported recently [100]. A number of effective USP2 inhibitors were identified using this assay. Natural substrate UBA52 was used and quantitative measurement was based on the infrared emission of Coomassie dye on SDS-PAGE.
This method uses readily available and inexpensive materials and has excellent reproducibility without the interference problem that is intrinsic to any fluorescence-based approaches. It also has the advantage of using a natural protein substrate, avoiding any artifacts that may be introduced by unnatural substrates. However, this assay was not amenable to high-throughput screening. It is useful for the accurate determination of IC50 values during fine-tuning of the structures during the structure-activity studies.
3.6. MADAL-TOF
A sensitive and fast assay to quantify in vitro DUBs enzyme activities using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry has been developed [101]. This method realized the high specificity of many members of the OTU and JAB/MPN/Mov34 metalloenzyme DUB families. It used unmodified substrates, such as di-ubiquitin topoisomers, and can be used to assess the potency and specificity of deubiquitylation inhibitors.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22094546
This entry is offline, you can click here to edit this entry!
