WUSCHEL-related homeobox (WOX) transcription factors (TFs) are well known for their role in plant development but are rarely studied in citrus. CsRAP2.12 and CsHB22 bind to the CsWUS promoter and regulate its activity. CsCYCD3 protein involved in cell proliferation interact with CsWUS protein
- Citrus sinensis
- CsWOXs
- transcription factor
- CsHB22
- CsCYCD3
- CsRAP2.12
1. Introduction
Homeobox transcription factors (TFs) containing homeodomain proteins are separated into 14 families, including the WUSCHEL -related homeobox (WOX) TF family [1,2,3]. The domain features a helix–loop–helix–turn–helix (HTH) structure that contains 60–66 amino acids crucial for specific functions in plants [4,5,6]. Two alpha-helices are intricately associated with DNA by a short turn, defined as the HTH motif [7]. WOX proteins usually contain a strongly conserved homeodomain that is highly specific to DNA binding proteins and may act as repressors or activators [3,8]. Homeodomain proteins are widely identified in monocot and dicot plants [9]. The model plant Arabidopsis contains 15 WOX members, which are divided into three clades based on their evolutionary relationship: modern/WUS, intermediate, and ancient clades [9]. Arabidopsis WOX members have been extensively studied, and their orthologs have exhibited diverse functions in numerous development processes [10,11,12,13], along with rice and maize [8,12]. Besides Arabidopsis, genome-wide studies have also been conducted on the WOX gene families of woody plants such as walnut [10], physic nut [14], grapes [15,16], peach, pear, apricot [17], coffee [18], and poplar [19]. In these plants, WOX genes are involved in vital regulatory networks that link the developmental mechanisms in plants, including shoot apical meristem, lateral organ development, plant stem cell maintenance, and floral determinacy [5,20,21,22].
In Arabidopsis, AtWUS and AtWOX1 determine floral meristem identity and maintenance [23,24,25]. AtWOX2, AtWOX6, AtWOX8, and AtWOX9 regulate ovule development and enable the development of cotyledon boundaries, eggs, and zygotes [26]. AtWOX3 targets pathways that promote flower organ primordia and leaf margin development [27]. AtWOX7 plays an important role in lateral root development and sugar (sucrose and glucose) status in Arabidopsis [28]. AtWOX11/AtWOX12 is involved in cell fate transition and root organogenesis [29]. Some WOX genes in Arabidopsis are involved in hormone signaling transduction pathways. For example, AtWOX4, AtWOX5, and AtWOX11 regulate auxin signaling that determines lateral organ and apical root growth [19,20,30]. WOX1 homologs have been demonstrated to control leaf blade outgrowth in Zea mays, Petunia hybrida, Medicago truncatula, and Nicotiana tabacum [5,31,32]. Leaf blade outgrowth is controlled by the WOX domain [33]. Recently, 12 WOX proteins were identified in walnut; JrWOX3a and JrWOX3b enable leaf development. PpWUS and PpWOX5 regulate embryo development in Pinus pinaster [3,10]. In Norway spruce, PaWOX3 promotes lateral organ outgrowth in conifers [34]. In recent years, several previous studies reported that WOX genes respond to abiotic stresses and hormone treatment, including Oraza sativa, Gossypium, and Brassica napus [8,23,35,36,37]. However, the response of WOX genes during abiotic stress has not been studied adequately in citrus.
Several previous studies indicated that the WUS gene is one of the most important genes in the WOX gene family and involved in numerous important developmental processes including size of shoot meristem, somatic embryo, as well as adventitious shoot and lateral leaf formation [5,38,39]. For example, AtWUS is crucial for shoot apical meristem maintenance to replace the function of WOX1 and PRESSED FLOWER (PRS) [5]. The ectopic overexpression of AtWUS in tobacco is involved in stem cell fate and lateral leaf formation [40]. In Medicago truncatula, the AtWUS homolog HEADLESS (HDL) is also involved in leaf development [31]. WUS gene functions downstream of the CLAVATA3 (CLV3) signaling pathway [41]. The WUS gene is expressed in the organizing center and enhances CLV3 expression in stem cells. Likewise, CLV3 negatively regulates meristem size by suppressing WUS expression [42,43]. WUS interacts with CYCLOIDEA 2 (CYC2) and regulates reproductive organ development (ovary, stigma, and style) in Chrysanthemum morifolium [44]. In the embryonic columella, WOX5 and CYCD3;3/CYCD1;1 facilitate cell proliferation, and CYCD3 plays a major role in normal cell division [45]. The WUS orthologs STERILE AND REDUCE TILLERING (MOC3/SRT1) and TILLERS ABSENT1/MONOCULM 3 are involved in bud formation and female fertility of rice [46]. The WUS gene regulates histone acetylation and interferes with HISTONE DEACETYLASE (HDAC) activity, which stimulates the auxin signaling pathway in stem cells [38]. In addition, WUS expression is indirectly repressed by AGMOUS (AG) and stimulates expression of the zinc finger TF C2H2 type (KNUCKLES), which in turn suppresses WUS expression directly or indirectly involved in the maintenance of floral meristem cells [5]. Thus far, functional characterization of WUS genes has been studied in other plants but is rarely studied in citrus.
The above-mentioned studies report that WOX TFs primarily affect plant development by regulating the expression of downstream genes. Notably, WOX protein cloning and functional analyses were predominantly focused on model plants such as Arabidopsis. However, there have been relatively few studies on the regulatory and genetic development role of the WOX family in citrus. The availability of the citrus genome database gives us a valuable genetic resource to study specific sweet orange genes [47,48]. A thorough screening of the citrus database allowed us to find the evolutionary, regulatory, and developmental role of WOX orthologs in sweet orange. In the current study, we identified 11 putative WOX members in Citrus sinensis. Tissue-specific expression patterns and co-expression profiles of CsWOXs under water deficit and floral inductive conditions were comprehensively studied. The subcellular localization, transactivation activity, and DNA-binding ability confirmed that CsWUS may be a TF. Moreover, CsWUS overexpression and the virus-induced gene silencing (VIGS) assay revealed new insights into floral organ development, stem cell activity, and leaf development in citrus. In addition, yeast two-hybrid assays and DNA-protein interactions confirmed the complex involvement of CsWUS in developmental regulatory networks.
2. Genome-Wide Identification and In Silico Subcellular Localization Prediction of CsWOX Gene Family
To identify citrus WOX genes, Arabidopsis and rice WOX proteins were used as queries and all resulting sequences were retrieved from the sweet orange database (http://citrus.hzau.edu.cn/cgi-bin/orange/blast) using BLASTP. After removing sequence redundancies of the same protein, a total of 11 potential WOX proteins were identified as being allied with CsWOX proteins (Table 1). We further confirmed that all of these CsWOX proteins contained the homeodomain (PF00046 and SM00389). The 11 putative WOX proteins corresponding to the gene were named according to their physical location (from top to bottom) on chromosomes 1–8 (Table 1). Notably, one gene (CsWOX10) is located on an unknown chromosome. The coding sequence (CDS) length of CsWOX genes varied from 582 bp (CsWOX7) to 1104 bp (CsWOX2), encoding polypeptides of 193–367 amino acids in length, with a predicted molecular weight range of 15,437.5–40,473.1 Da and a theoretical isoelectric point (pI) ranging from 5.4 to 11.5 (Table 2). In addition, the subcellular localization of CsWOX proteins was predicted (Table 2). The 3D structure of proteins was also predicted (Supplementary Figure S1). The predicted locations of three CsWOX proteins (CsWUS, CsWOX3, and CsWOX5) were found to be nuclear localized. The remaining members of CsWOX were projected to be localized in chloroplast, mitochondria, cytoplasm, or plastid.
Table 1. Characteristics of Citrus sinensis WOX genes.
| Name | Genome ID | Chromosome | Start Site | End Site | CDS bp | Protein Length (aa) |
|---|---|---|---|---|---|---|
| CsWUS | Cs1g25270 | Chr1 | 27428553 | 27430267 | 876 | 291 |
| CsWOX1 | Cs1g26550 | Chr1 | 28546972 | 28548579 | 840 | 289 |
| CsWOX2 | Cs2g05310 | Chr2 | 2845364 | 2848185 | 1104 | 367 |
| CsWOX3 | Cs2g16790 | Chr2 | 13618391 | 13620633 | 1011 | 336 |
| CsWOX4 | Cs3g23280 | Chr3 | 25600272 | 25601376 | 654 | 217 |
| CsWOX5 | Cs3g27390 | Chr3 | 28411631 | 28414854 | 701 | 233 |
| CsWOX6 | Cs5g27430 | Chr5 | 30010095 | 30011889 | 807 | 268 |
| CsWOX7 | Cs7g31470 | Chr7 | 31300824 | 31301506 | 582 | 193 |
| CsWOX8 | Cs8g17610 | Chr8 | 20428816 | 20430708 | 627 | 208 |
| CsWOX9 | Cs8g18280 | Chr8 | 20929316 | 20930953 | 747 | 248 |
| CsWOX10 | orange1.1t00075 | ChrUn | 1445775 | 1448402 | 1059 | 352 |
Table 2. Protein composition and physiochemical characteristics of CsWOX proteins.
| Name | GRAVY | Aliphatic Index |
Major Amino Acids Content [49] |
Predicted Localization | Instability Index |
MW (Da) | pI |
|---|---|---|---|---|---|---|---|
| CsWUS | –0.958 | 44.95 | S (14%), G (11%), N (7.9%) | nucl | 47.95 | 31,866.59 | 6.66 |
| CsWOX1 | –0.731 | 70.61 | Q (8.6%), S (8.2), L (8.6%) | nucl, chlo | 47 | 30,792.23 | 6.26 |
| CsWOX2 | –0.52 | 70.87 | Q (8.4%), S (12%), P (7.9%) | chlo, nucl, cyto_nucl, mito | 62.15 | 40,473.19 | 6.76 |
| CsWOX3 | –1.359 | 39.49 | N (14.0%), S (13%), T (11%) | Nucl | 46.22 | 26,406.6 | 10.21 |
| CsWOX4 | –1.136 | 37.54 | S (13%), T (10%), R (13.8%) | nucl, mito, cyto_nucl, extr | 52.15 | 15,437.5 | 11.5 |
| CsWOX5 | –0.811 | 59.44 | Q (10.3%), S (6%), A (6%) | nucl | 52.52 | 26,715.1 | 5.46 |
| CsWOX6 | –0.371 | 67.31 | S (11.9%), G (8.2%), A (7.5%) |
nucl, chlo, cyto | 61.21 | 29,123.41 | 5.61 |
| CsWOX7 | –0.927 | 59.53 | S (7.8%), T (6.7%), G (6.2%) | nucl, chlo, cyto | 50.27 | 22,461.97 | 6.25 |
| CsWOX8 | –0.756 | 60.48 | Q (11.1%), S (7.2%), L (8.7%) | nucl, cyto, mito, plas | 68.12 | 24,103.7 | 9.33 |
| CsWOX9 | –0.775 | 55.48 | G (8.5%), S (7.3%), Q (7.3%) | nucl, cyto, extr | 59.76 | 27,672.89 | 6.3 |
| CsWOX10 | –0.958 | 50.85 | S (10.2%), N (13%), G (9.3%) | nucl, cyto | 36.56 | 26,748.22 | 10.03 |
3. Phylogenetic Analysis and Gene Structure of CsWOX Genes
3.1. Phylogenetic Analysis of CsWOX Genes
To explore the phylogenetic relationship of WOXs between citrus and other model plants, a phylogenetic tree was resolved using 39 WOX family members from Arabidopsis (15 genes), rice (13 genes), and sweet orange (11 genes). The phylogenetic distribution showed that all CsWOX members were grouped into three clades, modern/WUS, intermediate, and ancient, consistent with previous WOX family distribution schemes [2]. The modern/WUS clade was the largest in this phylogenetic tree, containing 18 members: four members from sweet orange, six from rice, and eight from Arabidopsis. The intermediate clade was the second largest and included two from sweet orange, six from rice, and four from Arabidopsis members. The ancient clade had five members from sweet orange, one from rice, and three from Arabidopsis. Additionally, we also explored the orthologous relationships among sweet orange, rice, and Arabidopsis WOX families. These included 13 orthologous genes, putative orthologs with sweet orange proposed based on the phylogenetic tree were as follows: CsWOX5/AtWOX10, CsWOX1/AtWOX13, CsWOX6/AtWOX11, CsWOX2/AtWOX9, CsWOX8/AtWOX1, CsWOX7/AtWOX5, CsWOX9/AtWOX2, CsWOX1/OsWOX9B, CsWOX6/OsWOX11/12, CsWOX2/OsWOX9D, CsWOX8/OsNS1/2, CsWOX7/OsWOX5, and CsWOX9/OsWOX3 (Figure 1A).
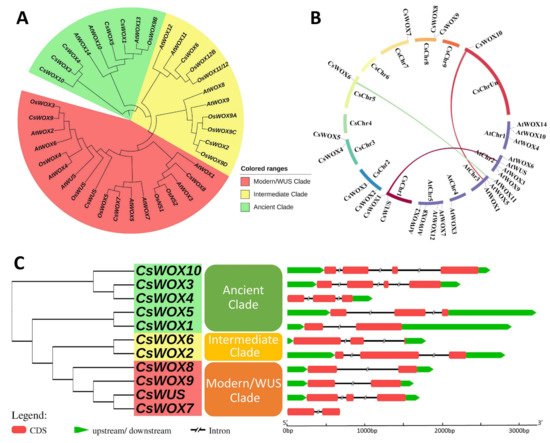
Figure 1. Phylogenetic tree, synteny analysis, exon, and intron distribution of WOX genes of citrus. (A) Phylogenetic tree of citrus CsWOX; rice (OsWUS: AM234746; OsWOX3: AM234749; OsWOX4: AM234750; OsWOX5: AM234751: OsWOX9A: Q0JKK6; OsWOX9B: AM234755; OsWOX9C: AM234752; OsWOX9D: AM234753; OsWOX11/12: AM234754; OsWOX12B: ABF95709; OsNS2: AM234748; OsNS1: AB218893); and Arabidopsis (AtWUS: AT2G17950; AtWOX1: AT3G18010; AtWOX2: AT5G59340; AtWOX3: AT2G28610; AtWOX4 AT1G46480; AtWOX5: AT3G11260; AtWOX6: AT2G01500; AtWOX7: AT5G05770; AtWOX8: AT5G45980; AtWOX9: AT2G33880; AtWOX10: AT1G20710; AtWOX11: AT3G03660; AtWOX12: AT5G17810; AtWOX13: AT4G35550; AtWOX14: AT1G20700). (B) Synteny analysis and chromosomal distribution of CsWOX genes; colored bars joining two chromosomal regions represents syntenic regions. Chr, Chromosome. (C) Exon-intron distribution. CDS exon indicated by red boxes, upstream and downstream region indicated by green boxes, intron indicated by black line.
3.2. Gene Structure and Synteny Analysis of CsWOX Genes
To further observe the structural diversity of the WOX genes in sweet orange, an exon–intron diagram of the CsWOX genes was created with reference to their genomic and coding sequences; the number of exons varied from two to four and the number of introns from one to three in CsWOXs (Figure 1C). Furthermore, MCScan was used to identify duplicate gene types (Figure 1B). Almost all CsWOX genes were singletons. In a syntenic block (Citrus sinensis and Arabidopsis), each member belonged to the same subfamily and phylogenetic group.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22094919
