Flowering plants develop new organs throughout their life cycle. The vegetative shoot apical meristem (SAM) generates leaf whorls, branches and stems, whereas the reproductive SAM, called the inflorescence meristem (IM), forms florets arranged on a stem or an axis. In cereal crops, the inflorescence producing grains from fertilized florets makes the major yield contribution, which is determined by the numbers and structures of branches, spikelets and florets within the inflorescence. The developmental progression largely depends on the activity of IM. The proper regulations of IM size, specification and termination are outcomes of complex interactions between promoting and restricting factors/signals.
- inflorescence meristem
- inflorescence architecture
- shoot apical meristem
- spikelet
- branching
- cereal crops
- yield improvement
1. Introduction
Higher flowering plants display a huge diversity of inflorescence architectures ranging from a solitary flower to specialized structures that contain multiple branches and florets. This variation is temporally and spatially controlled by two opposite activities: the maintenance of meristems for new lateral organs and determinacy of meristems during the flower formation. The inflorescence meristem (IM), which derives from the shoot apical meristem (SAM), produces organs on its flanks while maintaining a pool of pluripotent stem cells at its apex [1,2]. The inflorescence of dicot plant Arabidopsis thaliana exhibits an indeterminate pattern, in which the IM constantly produces secondary inflorescence meristems, as well as floral meristems (FMs) [3]. In the grass family (Poaceae), inflorescence architecture is largely established by iterations of branching. The basal unit of grass inflorescence is referred to as a spikelet, which is a short and condensed branch containing leaf-like structures. Specifically, after the SAM converts into IM, the IM initiates branch meristems (BMs), and/or spikelet meristems (SMs), which further initiate FMs [4,5]. Modeling inflorescence meristems have been proposed as transitioning from an indeterminate IM to a determinate FM [4]. Consequently, the variation of this progression creates a wide range of inflorescence types exist among closely related species, such as panicle, raceme, spadix, and spike [4,6]. Some grass inflorescences, such as the rice panicle and maize tassel, consist of a main axis, long branches and spikelets, but others, like wheat and barley belonging to the Triticeae tribe, characteristically show an unbranched spike-type inflorescence that spikelets are directly attached to the inflorescence axis. The variation in inflorescence architecture is controlled by the activity of specialized meristems: the branch meristem and the spikelet meristem [6].
It is recognized that meristem determinacy is key to understanding inflorescence architecture [4]. The genetic basis of inflorescence initiation and development has been extensively studied in the eudicot Arabidopsis and monocot crops such as rice and maize [2,6]. For example, classic Arabidopsis CLV (CLAVATA)-WUS(WUSCHEL) negative regulatory loop controls SAM activity and size, and further affects IM formation and differentiation [1]; in rice and maize, regulatory mechanisms of conserved CLV signaling underlying meristem size control and inflorescence specification, have also been demonstrated [7]. Additionally, a large collection of grass genes and domestication QTL (quantitative trait loci) associated with inflorescence architecture has been identified [8,9,10,11,12,13,14,15]. The reported regulators are involved in peptide-receptor signaling, G protein pathway, plant hormone pathway, photoperiod signaling, transcription factor regulatory networks and microRNA-targets modules, which provide key insights into the molecular mechanisms regulating inflorescence architecture. Mutations of these players in grasses lead to the altered IM determinacy and size, resulting in the changed flower/spikelet number, inflorescence architecture, and final grain yield [1,3,5,6,7,16]. Importantly, these orthologous regulators and pathways among cereals show functional conservation and divergence in regulating inflorescence development and yield components.
Enhancing the yield potential and stability of main cereals that are utilized as staple food and feed for humans and livestock, such as rice, maize, wheat, and barley, is a priority for global food security [17]. Crop selection and breeding have generated variants with increased yield through optimizing inflorescence traits, such as branching and spikelet number. Investigations of genes/alleles associated with inflorescence architecture have revealed basic developmental patterning mechanisms, which are pivotal for genetic approaches to optimize yield in cereal crops [12,13,14,15,18]. The role of spikelet development in yield improvement has been well-reviewed recently [19,20]. Genetic and genomic advances in cereal species have uncovered the molecular determinants of inflorescence architecture and facilitated knowledge transferring across genera.
2. The Structure and Developmental Fate of IM in Cereal Crops
The meristems of higher plants are centers of cell proliferation and organ initiation. Morphological studies have revealed that the SAM comprises a central zone of slowly dividing, undifferentiated cells and a peripheral zone of more rapidly dividing cells that are in transition towards specification [3]. In grass family, upon the transition from vegetative phase to reproductive phase, SAM ceases producing leaves on its flanks and turns into IM that, depending on the species, starts producing lateral branch meristems (BMs) or directly spikelet meristems (SMs). Therefore, neither the IM nor the BMs are directly converted to floret meristems (FMs). Instead, all the higher-order meristems produced by the IM and its branches are ultimately converted to SMs, which first produce two bracts known as glumes, followed by one or more FMs in each spikelet. Because the development of the spikelet is highly stereotyped and deterministic within most major groups of grasses, the spikelet is defined as the terminal differentiated unit of the inflorescence, rather than florets [4,6]. In other words, the IM produces either BMs or SMs on its flanks, and the BMs themselves further produce either secondary BMs or SMs. During this progression, different sets of developmental decisions lead to the diversity of inflorescence architecture in grasses [21,22].
Rice has a panicle-like inflorescence that gives rise to primary and secondary branch meristems (pBMs and sBMs) and further generates SMs and FMs, respectively, in a determinate pattern (Figure 1), producing an average of 150–250 grains per inflorescence in modern cultivars. More complicatedly, maize has two distinct inflorescences, tassel and ear, bearing male and female flowers, respectively. The apical tassel is derived from the SAM and consists of indeterminate two-rowed branches at its base and a many-rowed central spike, whereas an ear is positioned laterally in the axils of leaves and consists of a many-rowed axis bearing spikelets (Figure 1). Both tassel and ear have determinate paired SMs, each initiating two FMs [4,6,22]. One of the two pistillate florets in each spikelet of the ear is fertile and the other is aborted, and an ear is able to produce an average of 350–450 grains (Figure 1). In grass tribe Triticeae, like barley and wheat, inflorescences exhibit a branchless spike architecture with spikelets directly attached to the axis. In barley, the IM differentiates into many spikelet ridges and each one forms a final triple spikelet meristem (TSM) carrying three individual SMs [4,5]. The triple spikelet is composed of one central spikelet and two lateral spikelets, and the fertility of lateral spikelets is either suppressed to form a two-rowed inflorescence type with 20–35 grains per spike or promoted to form a six-rowed type carrying average 45–80 grains per spike (Figure 1). In wheat, the IM differentiates into axillary meristem (AM) that includes a lower leaf ridge and upper spikelet ridge. The spikelet ridge differentiates into an SM that forms several FMs in a distichous manner on the indeterminate rachilla [18]. Ultimately, each wheat spike produces average 50–80 kernels in most of modern cultivars. Importantly, distinct fates of meristems are decided by specific genetic control, molecular regulation and extrinsic environmental signals, leading to a great plasticity of grass inflorescences and yield variation.
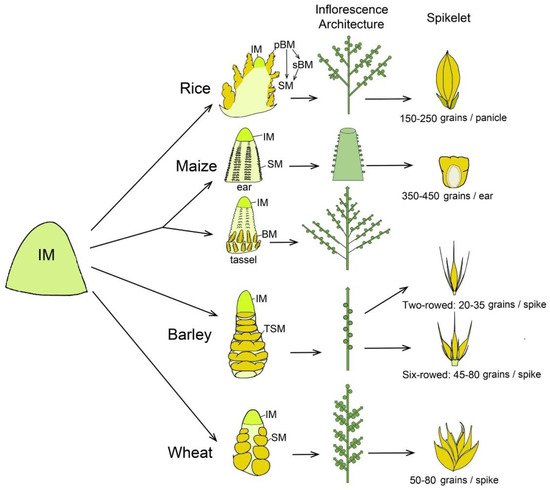
Figure 1. Schematic representation to compare inflorescence meristem differentiated fate, inflorescence architecture and spikelet in rice, maize, barley and wheat. In rice, inflorescence meristem (IM) generates two types of lateral branch meristems (BMs). The primary branch meristems (pBMs) generate spikelet meristems (SMs) and secondary branch meristems (sBMs). The sBMs further produce more SMs. In maize, IM of tassel is converted from shoot apical meristem, while the axillary meristem converts into an ear. The ear IM initiates a series of determinate axillary meristems, giving rise to pairs of SMs. The tassel produces BMs, which then form pairs of SMs. Each SM of ear and tassel further initiates two floret meristems (FMs). In barley and wheat, the IM directly differentiates SMs in axis without forming BMs. Barley has a triple spikelet meristem (TSM) structure composed of a central spikelet and two lateral spikelets, whose development is either suppressed to form a two-rowed type or promoted to form a six-rowed type. Conversely, in wheat, the inflorescence is composed of single spikelet that produce multiple FMs.
3. The Genetic Regulation of Transition from SAM to IM
3.1. CLV Pathway
The SAM is in a balance between providing founder cells for new organs and maintaining its own stem cell niche. Manipulating the meristem size or breaking this balance could induce plants to produce more organs. The CLV-WUS negative feedback loop regulates the stem cell maintenance, which has been adequately investigated in Arabidopsis. The WUS gene, encoding a homeodomain transcription factor, induces expression of the CLV3 gene. The CLV3 is a secreted peptide and is perceived by receptor complexes including leucine-rich repeats (LRRs) kinases CLV1 and CLV2 that, upon activation, stabilize the meristem stem cell population by signaling back to repress the expression of WUS, thereby completing the negative feedback loop [1,7]. Mutations of CLV1, CLV2 and CLV3 show increased IM size, resulting in increased numbers of flowers and floral organs [23,24,25].
The CLV signaling pathway is basically conserved in grass species (Figure 2 and Table 1) [7,26,27,28,29]. In rice, FLORAL ORGAN NUMBER 1 (FON1) gene encodes a leucine-rich repeat receptor-like kinase similar to CLV1 in Arabidopsis [26]. FON2 (also called FON4) is an ortholog of Arabidopsis CLV3 [28,30]. The fon2/4 mutants exhibit an increased size of SAMs and FMs, and then increased number of both primary branches and floral organs. By contrast, overexpression of FON2 leads to a smaller IM with reduced floral organs, indicating that FON2 has similar roles of CLV3 in rice. Genetic analysis of fon1 fon2 double mutants suggests that FON1 and FON2 function in the same genetic pathway and FON2 may be the ligand of FON1 [30], indicating the conserved function of CLVs in limiting meristem size. Moreover, another two CLV3-like genes in rice, FON2-LIKE CLEPROTEIN1 (FCP1) and FCP2, show similar expression patterns with broad accumulations in the SAM. Genetically, FCP1 and FCP2 function redundantly to maintain SAM activity independently from FON1 [31]. Notably, the interaction between FON2 and the rice ortholog of WUS, named TILLERS ABSENT1 (TAB1; also called OsWUS), happens in a slightly different scenario where TAB1 is only required to maintain stem cells during axillary meristem development with FON2 restricting its expression [32]. It is likely that the FON2-TAB1 pathway has been recruited to play a specific role within a narrow developmental window in rice during evolution.
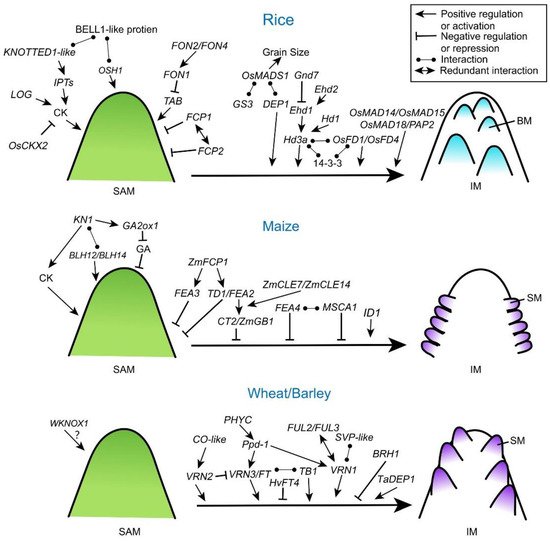
Table 1. Key regulators involved in the transition from SAM to IM in rice, maize, barley and wheat.
|
Rice |
Maize |
Barley |
Wheat |
Pathways |
Reference |
|---|---|---|---|---|---|
|
FON2/4 |
ZmCLE7; ZmCLE14 |
CLV-WUS |
|||
|
FCP1; FCP2 |
ZmFCP1 |
CLV-WUS |
|||
|
FON1 |
TD1 |
CLV-WUS |
|||
|
FEA2 |
CLV-WUS |
[33] |
|||
|
FEA3 |
CLV-WUS |
[34] |
|||
|
TAB1 |
CLV-WUS |
[32] |
|||
|
OSH1 |
KN1 |
WKNOX1 |
KNOX |
||
|
BLH12; BLH14 |
KNOX |
[43] |
|||
|
CT2 |
G-protein |
||||
|
ZmGB1 |
G-protein |
[56] |
|||
|
GS3 |
TaDEP1 |
G-protein |
|||
|
Maize Gα |
Brh1 |
G-protein |
|||
|
Hd3a |
HvFT1; HvTF2; HvFT4 |
VRN3; FT1; FT2 |
Photoperiod |
||
|
Ehd1 |
Photoperiod |
||||
|
Hd1 |
HvCO1; HvCO2 |
Photoperiod |
|||
|
Ghd7 |
ZmCCT10 |
HvVRN2 |
TaVRN2 |
Photoperiod |
|
|
OsFD1; OsFD4 |
Photoperiod |
||||
|
Ehd2 |
ID1 |
Photoperiod |
|||
|
Ppd-H1 |
Ppd-1 |
Photoperiod |
|||
|
OsMADS1; OsMADS14; OsMADS15; OsMADS18 |
HvVRN1 |
FUL2; FUL3; TaVRN1 |
Others |
||
|
FEA4 |
Others |
[87] |
|||
|
MSCA1 |
Others |
[88] |
|||
|
OsCKX2; LOG |
HvCKXs |
TaCKXs |
Others |
Abbreviations of gene names: FON, FLORAL ORGAN NUMBER; CLE, CLAVATA3/ESR-related; FCP, FON2-LIKE CLE PROTEIN; TD1, THICK TASSEL DWARF1; FEA, FASCIATED EAR; TAB1, TILLERS ABSENT1; OSH1, Oryza sativa HOMEOBOX 1; KN1, KNOTTED1; BLH, BELL1-like homeobox protein; CT2, COMPACT PLANT2; ZmGB1, Zea mays Gβ subnuit; GS3, Grain Size 3; DEP1, DENSE ERECT PANICLE1; Brh1, Brachytic1; Hd3a, Heading date 3a; FT, FLOWERING LOCUS T; VRN, VERNALIZATION; Ehd1, Early heading date 1; Hd1, Heading date 1; CO, CONSTANS; ZmCCT10, CO, CONSTANS, CO-LIKE and TIMING OF CAB1; Ghd7, Grain number, plant height, and heading date 7; FD, FLOWERING LOCUS D; ID1, INDETERMINATE 1; Ppd, Photoperiod-H1; FUL, FRUITFULL; MSCA1, MALE STERILE CONVERTED ANTHER1; CKX, Cytokinin Oxidase/dehydrogenase; LOG, LONELY GUY.
In maize, THICK TASSEL DWARF1 (TD1) and FASCIATED EAR2 (FEA2) genes are the closest orthologs of Arabidopsis CLV1 and CLV2, respectively [27,33]. Mutations in these genes result in extensive overproliferation of stem cells and meristem fasciation, and the td1 fea2 double mutant has more severe phenotypes than the single mutants [27]. In addition, ZmCLE7 (CLAVATA3/ESR-related peptide) and ZmCLE14, have been identified as potential CLV3 orthologs and both of them have a negative effect on SAM size [34].
Another maize CLV3-like gene, ZmFCP1, an ortholog to rice FCP1, also plays a key role in meristem specification, and zmfcp1 mutants show a clv-like ‘fasciated ear’ phenotype. However, ZmFCP1 and ZmCLE7 signals are transmitted by FEA2 through different protein complex [35], suggesting that these CLV3-like peptides can parallelly regulate stem cell homeostasis in meristems. A new maize CLV receptor, FEA3, similar to FEA2 in sequence, has been reported to integrate into a mathematic model of the CLV–WUS feedback [34]. The fea3 mutants show enlarged and fasciated meristems, and are partially insensitive to ZmFCP1 peptide treatment [34]. Although fea2 mutants do not increase overall yield due to a compensatory reduction in kernel size, a decrease of FEA2 expression level can increase IM size as well as kernel row number [36]. Recently, a weak allele of FEA2 has been found having higher kernel row number and yield in maize [37]. FEA3 acts in a separate pathway than FEA2 but, similarly, the weak allele of FEA3 increases kernel row number [34]. Therefore, the multiple-member CLV pathway has the potential to enhance grain yield in maize.
Although removing CLVs restrictions on meristems produces more spikelets, this effect is more likely due to increases of organs number rather than a prolonged indeterminate status of meristems, because higher order branches are rarely formed in maize ears of these mutants. Even though some CLV-like and WUS-like genes are highly expressed in developing barley inflorescence [38], the effects of the CLV pathway in wheat and barley inflorescence development are less reported. In wheat genome, 104 CLV3/CLE peptides have been identified. Phylogenetic analysis and chemically synthesized peptides treatment have revealed that these CLV3/CLEs may have distinct roles in regulation of root and shoot development [39]. All orthologs of CLVs and WUS in wheat and barley still remain unknown because of the lack of mutant resources. Future studies are required to explore CLV pathways and assess their contributions to grain yield in Triticeae crops.
3.2. KNOTTED 1-Like Homeobox (KNOX) Proteins
Class I KNOTTED 1-like homeobox (KNOX) proteins are key homeodomain transcription factors that regulate shoot apical meristem establishment and maintenance in plants [3,6]. In Arabidopsis, SHOOT MERISTEMLESS (STM) encodes a KNOTTED1 (KN1)-related homeodomain protein and stm mutants show very severe phenotype lacking the shoot meristem, which is fatal to seedlings [40]. Recent studies have uncovered that STM integrates into WUS-CLV loop to modulate the stem cell homeostasis [41]. In grasses, KNOX proteins are also required for the maintenance of the proper size and activity of the stem cell niche (Figure 2 and Table 1). Loss of function of KN1 (KNOTTED 1) in maize leads to the defects of shoot meristem maintenance and inflorescence development [42]. Two maize BELL1-like homeobox (BLH) transcription factors, BLH12 and BLH14, act as cofactors of KN1 and accumulate in overlapping domains in shoot meristems. Similar to kn1 mutants, blh12 blh14 double mutants fail to maintain axillary meristems and develop abnormal tassels [43]. Consistently, mutation of Oryza sativa HOMEOBOX 1 (OSH1) in rice, the maize KN1 ortholog, shows abnormal SAM, smaller inflorescences and a decreased number of spikelets [44]. Rice BELL1-like homeobox genes are also involved in regulating inflorescence architecture and meristem maintenance [45]. Similarly, rice BELL1-type proteins form a heterodimer with KNOX-type proteins such as OSH1 [46]. It is likely that heterodimers of KNOX-type and BELL1-like proteins play a crucial role in maintaining the SAM in grasses. In wheat, the KN1-like homeobox gene, WKNOX1, is expressed in SAM-containing shoots and young spikes [47], but its genetic function is still unknown.
Exploring KN1 downstream targets is a potential way to understand the mechanisms of KNOX proteins in meristem maintenance. In maize, KN1 coordinates the regulatory gene networks by directly binding to a huge number of loci and genes, most of which encode key regulators involved in auxin, cytokinin, and gibberellic acid (GA) signaling pathways [48]. Maize KN1 represses the accumulation of bioactive GA directly through positive regulation of GA2ox1 (Gibberellin 2-beta-dioxygenase 1) that encodes a GA-inactivating enzyme, which is required for proper establishment and maintenance of SAM and IM [49]. In rice, ectopic expression of KNOTTED1-like homeobox protein promotes the accumulation of cytokinin by activating cytokinin biosynthesis genes adenosine phosphate isopentenyltransferases (IPTs) [50]. Therefore, KNOX proteins may play a key role in regulating SAM activity by coordinating phytohormones in cereal plants (Figure 2), which is consistent with findings in dicot plants [51,52].
3.3. G-Protein Pathway
Heterotrimeric G proteins contain Gα, Gβ and Gγ subunits and play a critical role in signal transmission [53]. Compared to the classic heterotrimeric G protein signaling in the mammalian system, plant G proteins are less understood [53,54]. Recently, several G protein subunits have been demonstrated contributing to meristem specification and inflorescence architecture in cereal plants (Figure 2 and Table 1). A maize Gα protein COMPACT PLANT2 (CT2) functionally interacts with the CLV receptor FEA2 to control SAM development [35,36]. In ct2 loss-of-function mutants, SAM size is increased, and thicker tassels and denser ears are formed, which resembles Arabidopsis clv and maize fea2 mutants, indicating that CT2 transmits a stem-cell-restrictive signal from a CLV receptor in maize [36]. Expression of a constitutively active CT2 results in the increased size of ear IMs, and higher spikelet density and kernel row number, all beneficial traits selected during maize improvement [55]. Knock out mutation of maize Gβ subunit (ZmGB1) shows a lethal phenotype, and genetic screening for suppressor of the lethal phenotype has revealed that ZmGB1 acts with Gα subunit gene to control meristem size and activity, indicating that Gβ and Gα are in a common signaling complex in maize. Moreover, ZmGB1 functions together with CT2 in downstream of the FEA2 CLAVATA receptor during inflorescence development (Figure 2) [56]. However, the role of maize Gγ subunit in shoot meristem development remains unknown. In rice, a Gγ subunit, rice Dense and Erect Panicle 1 (DEP1), has been identified as a major QTL controlling panicle branching, seed size, and seed number, and wheat TaDEP1 also shows the similar function in regulating spike development [10,57]. Another rice G-protein γ subunit, Grain Size 3 (GS3), whose loss-of-function allele forms longer but fewer grains, acts the key regulator for grain shape and yield production. Supportively, natural variation in GS3 can explain about 79% of the variation between short-grain versus long-grain cultivars [9]. Both Gγ subunits GS3 and DEP1 interact directly with a conserved floral homeotic E-class MADS-box protein, OsMADS1, to regulate grain size and shape [58]. In barley, a semi-dwarfing gene, Brachytic1 (Brh1), encodes a G protein α subunit. Brh1 mutation causes a shorter spike and rounded grain shape [59]. Except maize, currently there is a lack of proofs for heterotrimeric G protein subunits from other cereal crops participating in the crosstalk with CLV pathways.
3.4. Genetically Controlled Photoperiod Response in Meristem Specification
The transition from SAM to IM in cereals is also controlled by orchestration and integration of endogenous signals and exogenous signals, such as temperature and photoperiod [5,6,18]. In molecular level, this transition is accomplished by the florigen activation complex (FAC) (Figure 2). For example, rice florigen FT (FLOWERING LOCUS T)-like protein Hd3a binds its cofactor 14-3-3 protein to form a complex which further recruits bZIP transcription factors, like OsFD1 (FLOWERING LOCUS D 1) and OsFD4, to promote the phase transition [60,61]. The photoperiod-dependent accumulation of florigen protein at apical meristem is critical for the activity of FAC. Hence, upstream regulators of florigen are directly or indirectly affecting apical meristem activity. Heading date 1 (Hd1), an upstream regulator of Hd3a, the rice ortholog of Arabidopsis photoperiod response regulator CONSTANS (CO), is an enhancer of phase transition under short-day conditions [8]. Rice Early heading date 1 (Ehd1) is a type-B response regulator and promotes floral transition under short-day conditions through upregulating Hd3a expression [62,63]. Grain number, plant height, and heading date 7 (Ghd7) acts as a floral repressor inhibiting Ehd1 in the context of phytochrome signals [64]. Monocot-specific zinc-finger transcription factors, Ehd2 of rice and INDETERMINATE 1 (ID1) of maize, also have been reported to regulate meristem activity by controlling photoperiodic pathway through Ehd1 [65,66]. Importantly, impacts of these photoperiodic regulators appear beyond the phase transition at apical meristem. Enhanced expression of Ghd7 increases the number of secondary branches and panicle size in rice under long-day conditions [67]. Overexpression of the maize photoperiod response gene, ZmCCT10 (CO, CONSTANS, CO-LIKE and TIMING OF CAB1), an ortholog of the rice Ghd7, modifies flowering time and inflorescence morphology [68]. Naturally occurring rice genetic alleles of Hd1, Ehd1, Ghd7 and Ghd8, have been used in breeding aiming to higher grain yield [11,63,67,69], yet how these photoperiodic genes regulate inflorescence meristem specification afterwards remains largely unknown.
In Triticeae, functionally homologous regulators of flowering have also been investigated (Figure 2). Wheat VRN3 (VERNALIZATION 3) gene encodes an ortholog of FT protein, and mutations of this gene in bread and tetraploid wheat delay the transition to reproductive growth, prolong the duration of spike development, and increases the number of spikelets [70,71]. Overexpression of a barley FT-like gene (HvFT4) specifically delayed spikelet initiation and reduced the number of spikelet primordia and grains per spike [72]. Significantly, both wheat and barley have a major QTL contributing to photoperiodic regulation of flowering called Photoperiod-1 (Ppd-1) [73,74]. Wheat Ppd-1 influences inflorescence architecture and paired spikelet development by modulating the expression of FT1 under short day condition [74]. Moreover, Ppd-1 activates FT2, and the latter regulates the number and formation of spikelets [75]. PHYTOCHROME C (PHYC) acts upstream activating Ppd-1 and FT1 in tetraploid wheat, and the phyc mutant delays flowering and alters spike development [76]. Additionally, wheat Teosinte Branched1 (TaTB1), a TCP (Teosinte branched1/Cincinnata/Proliferating cell factor) family transcription factor, is decoupled from photoperiod but can interact with the florigen protein, FT1, and regulate inflorescence branching [77]. In barley, a gene network closely associated to Ppd-1 has been investigated, including FT2, floral homeotic MADS-box genes like SEPALLATA1 (SEP1), SEP3, PISTILLATA (PI), APETALA3 (AP3), and VRN1 [78]. These genes are highly expressed in developing IMs [38]. Ppd-1 is also involved in the crosstalk with vernalization flowering pathway. Together with CO homologs in barley, HvCO1 and HvCO2, Ppd-1 represses flowering by up-regulating HvVRN2 (the ortholog of rice Ghd7 and wheat TaVRN2) expression before vernalization but promotes HvFT1 after vernalization [79,80]. During the phase transition of the apical meristem, major cereals share common regulatory patterns in response photoperiod. More importantly, the flowering regulators often continue playing an active role in the IM development, therefore contributing to yield production.
3.5. Other Pathways
As an outstanding family of regulators active throughout the entire reproductive development, MADS-box transcription factors are closely associated with the phase transition of grass inflorescences [6]. For instance, knockdown of three APETALA 1 (AP1)/FRUITFULL (FUL) family members, OsMADS14, OsMADS15, and OsMADS18, in a rice sepallata (SEP) mutant panicle phytomer 2 (pap2) background shows a delayed transition from SAM to IM and produce multiple shoots instead of one inflorescence (Table 1) [81]. As to Triticeae, VRN1, encoding a MADS box transcription factor, is induced by vernalization to trigger meristem transition and flowering in wheat and barley [82]. Ectopic expression of barley VRN1 protein accelerates the transition to reproduction and flowering [83]. Wheat TaVRN1 cooperates with another SVP (SHORT VEGETATIVE PHASE) -like MADS protein to regulate vernalization-induced reproductive transition [84]. In addition, wheat MADS-box genes, VRN1, FUL2 and FUL3 have redundant roles in regulation of the transitions from the vegetative SAM to IM and from IM to spikelet. The ful2 null mutant produces more florets per spikelet, additive to a higher number of spikelets, resulting in a significant increase in the number of grains per spike in the field [85]. Moreover, complexes of wheat FUL and SVP act during the early reproductive phase to promote heading and formation of the spikelet [86]. Thus, these FUL family genes are essential in the acquisition and termination of IM identity (Figure 2). Whether other MADS-box genes also contribute to the transition from SAM to IM in cereal crops needs further investigation.
Besides as components of FAC, bZIP transcription factors also regulate meristem activities via other pathways. Loss of function of maize FEA4 leads the increased meristem size and similar fascitated phenotype from maize fea2 and fea3 mutants [87]. FEA4 promotes expression of many genes involved in meristem determinacy and auxin signaling, which shares some targets with KN1. Therefore, FEA4 and KN1 may act antagonistically, in controlling the determinacy–differentiation balance [87]. FEA4 activity is controlled by a redox mechanism, via interacting with the glutaredoxin MALE STERILE CONVERTED ANTHER1 (MSCA1). Dominant mutations in MSCA1 show bigger meristems and loss-of-function mutants of msca1 correspondingly have smaller shoot meristems [88]. Redox signaling also play a key role in the controlling of shoot meristem size in other plants by regulating WUS expression [89].
Plant hormone cytokinin is essential in the regulation of meristematic activity, inflorescence branching in plants [1]. Mutations in rice OsCKX2 (a cytokinin oxidase/dehydrogenase) and LONELY GUY (LOG, a cytokinin-activation enzyme) could lead to altered cytokinin distribution in meristems and consequently change the SAM, IM and BM activities [90,91]. In barley, the dynamic of cytokinin is also required for inflorescence meristem maintenance and spike architecture [92]. Modification of cytokinin content via manipulating the key cytokinin oxidase/dehydrogenase genes has become a potential way for yield improvement in wheat and barley [93,94].
4. IM Differentiation: Branches or Spikelets
After the initiation of IM, patterns of determinacy in IM shape the inflorescence morphology. The developmental fate of the IM in grasses, i.e., its conversion into a SM or its production of branch meristems that convert to SMs later, is species specific and determines the architecture of inflorescence [4]. In rice, before completing the terminal SM development, the whole IM maintains an indeterminate status, generating primary and secondary branches, and forms a branching architecture (panicle) [6]. While meristem determinacy in wheat and barley is directly caused by the SM identity, which ultimately produces florets without branches. Wheat inflorescence is determinate and produces a terminal spikelet at the apex, whereas the barley inflorescence is indeterminate [5,18]. Therefore, IM determinacy and maintenance control the numbers of branches and spikelets. Multiple factors and pathways synergistically regulate the IM specification and activity (Figure 3 and Table 2), further affecting yield performance in cereal crops.
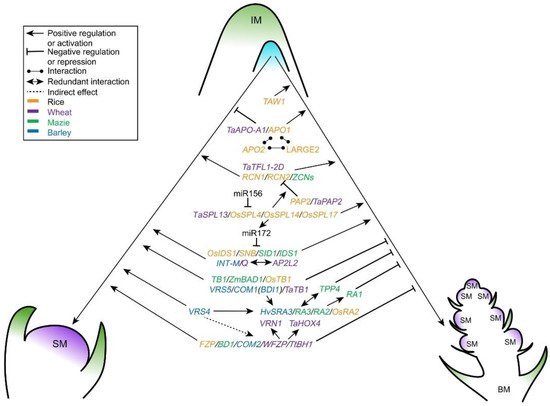
Figure 3. Genetic regulation of IM differentiation in cereal crops. The multiple players, including MADS, TCP, AP2 and SPL transcription factors, RA proteins, miRNAs, and their crosstalk genetically control BMs or SM identity in rice, maize, wheat and barley. IM, inflorescence meristem; BM, branch meristem; SM, spikelet meristem.
Table 2. Key regulators involved in IM differentiation and specification in rice, maize, barley and wheat.
|
Rice |
Maize |
Barley |
Wheat |
Pathways |
Reference |
|---|---|---|---|---|---|
|
PAP2/OsMADS34 |
TaPAP2 |
MADS-RCN |
|||
|
RCN1; RCN2 |
ZCNs |
TaTFL1 |
MADS-RCN |
||
|
RA1 |
RAMOSA |
[106] |
|||
|
OsRA2 |
RA2 |
VRS4 |
RAMOSA |
||
|
RA3 |
SRA3 |
RAMOSA |
|||
|
TPP4 |
RAMOSA |
[110] |
|||
|
FZP |
BD1 |
COM2 |
TtBH1; WFZP |
FZP |
|
|
OsTB1; OsTB2/REP1 |
TB1; ZmBAD1 |
VRS5; COM1/BDI1 |
TaTB1 |
TCP |
|
|
APO1; APO2 |
TaAPO-A1 |
Others |
|||
|
TAW1 |
Others |
[133] |
|||
|
OsSPL14; OsSPL4; OsSPL17 |
TaSPL13 |
Others |
|||
|
SNB; OsIDS1 |
IDS1; SID1 |
INT-M/DUB1 |
AP2L2; Q |
Others |
Abbreviations of gene names: PAP, PANICLE PHYTOMERL; RCN, MADS—RICE CENTRORADIALIS; ZCN, ZEA CENTRORADIALIS; TFL1, TERMINAL FLOWER1; RA, RAMOSA; VRS, Six-rowed spike; SRA3, SISTER OF RAMOSA3; TPP4, Trehalose-P-phosphatase 4; FZP, FRIZZY PANICLE; BD1, BRANCHED SILKLESS1; COM, COMPOSITUM; TtBH1, BRANCHED HEAD1; WFZP, wheat FZP; TB, TEOSINTE BRANCHED; REP1, RETARDED PALEA1; BAD1, BRANCH ANGLE DEFECTIVE 1; BDI1, BRANCHED AND INDETERMINATE SPIKELET 1; APO, ABERRANT PANICLE ORGANIZATION; TAW1, TAWAWA1; SPL, Squamosa Promoter Binding Like Protein; SNB, SUPERNUMERARY BRACT; IDS1, INDETERMINATE SPIKELET 1; SID1, Sister of INDETERMINATE SPIKELET 1; INT-M/DUB1, INTERMEDIUM-M/DOUBLE SEED1; AP2L2, APETALA 2-Like gene 2; Q, APETALA 2-Like gene 5.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22073508
