Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Plant Sciences
The plant hormone abscisic acid (ABA) rapidly accumulates in plants in response to environmental stress and plays a pivotal role in the reaction to various stimuli. Increasing evidence demonstrates a significant role of ubiquitination and subsequent selective degradation in controlling ABA signaling. We present updated information on the components of the ABA signaling pathway modified by different E3 ligases.
- abscisic acid
- ubiquitination
- E3 ubiquitin ligases
1. Introduction
Plants are often subjected to various detrimental environmental stressors. Despite their sessile lifestyle, plants are able to maintain their internal equilibrium when exposed to external challenges. This balance, called homeostasis, can be achieved by internal mechanisms that alter cell metabolism, thanks to which the whole organism can maintain relatively constant conditions. Such rapid and appropriate genetic and metabolic acclimation, which is a crucial issue for plants’ survival, is maintained due to the coordinated action of plant hormones and cellular degradation mechanisms influencing proteostasis.
Protein homeostasis (or proteostasis) integrates cellular pathways that mediate biogenesis, folding, trafficking, and the degradation of polypeptides to maintain the required concentrations of all proteins that compose the proteome. This field has been intensively investigated by numerous research groups [1]. Proteins are often tagged for removal by ubiquitination. Polyubiquitinated proteins and protein aggregates are degraded via the ubiquitin–proteasome system (UPS) pathway and via the autophagy–vacuolar route. The UPS pathway is restricted in its ability to degrade aggregated proteins, which are too large to pass through the narrow proteasome entrance channel [2]. Thus, autophagy represents a major mechanism in plants for degrading macromolecular ubiquitinated protein aggregates as a response to stressors [3][4].
Phytohormones are not only involved in protein degradation mechanisms, but also play a pivotal role in plant responses to various stimuli. Although the relationships between them are poorly understood, it is generally accepted that extensive crosstalk and multiple regulatory loops are responsible for maintaining the balance of growth and stress responses [5].
Phytohormones are master regulators of plant growth, development, and stress response. Of them, abscisic acid (ABA) is rapidly accumulated in plants in response to abiotic stress [6]. Similarly to most other hormones, ABA is involved in developmental processes, including seed maturation, seed dormancy and germination, primary root growth, and flowering time control, and in the response to adverse environmental stressors [6][7]. Plants exposed to abiotic stress quickly activate the ABA signaling cascade, resulting in the activation of ABA-responsive transcription factors and the induction of ABA-responding genes. However, after achieving stress tolerance, it is necessary to terminate (or attenuate) the ABA pathway. The balance is maintained by the selective degradation of specific components of the ABA signaling pathway upon their tagging for decay by ubiquitination. Conventional degradation of the tagged components takes place via the proteasome; however, increasing evidence demonstrates a significant role of autophagy in controlling ABA signaling [5][8][9][10][11][12]. In addition to this interplay with degradation systems, an additional layer of complexity is provided by extensive crosstalk between ABA and other phytohormones [13][14][15][16].
2. Principles of ABA Perception and Signaling and Their Control by Selective Degradation Systems
Reversible protein phosphorylation is a post-translational modification catalyzed by protein kinases and phosphatases. It acts as a molecular switch for almost all key events of cell metabolism and signaling in eukaryotes. Additionally, in the field of ABA signal transduction, protein kinases and phosphatases play pivotal roles, forming a reversible protein phosphorylation cascade, including ABA receptors (pyrabactin resistance/regulatory components of ABA receptor (PYR/RCAR)), type 2C protein phosphatases (PP2Cs), protein kinases (Snf1 (sucrose-non-fermentation 1)-related kinases subfamily 2, SnRK2s), and downstream targets. The key function of RCARs is to indirectly control the activity of SnRK2s, which phosphorylate numerous stress-activated targets in the response to ABA, including ABA-responsive transcription factors (TFs) [17][18]. This is accomplished through a negative regulatory pathway, involving RCARs, PP2C, and SnRK2 proteins and E3 ubiquitin ligases (Figure 1).
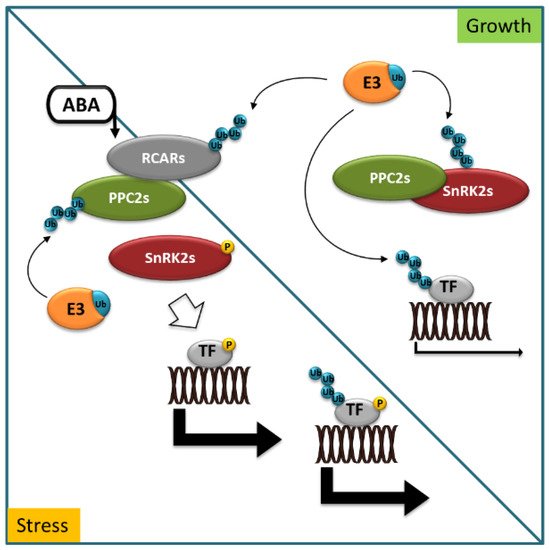
Figure 1. Abscisic acid (ABA) signaling in stress and growth. In growth-promoting conditions, ABA receptors (RCARs), protein kinases (SnRK2s) dephosphorylated by type 2C protein phosphatases (PP2Cs), and ABA-responsive transcription factors (TF) are marked for degradation with ubiquitin (Ub) by specific E3 ligases (E3). In stress conditions, ABA accumulates and binds to RCARs, allowing for interaction with PP2Cs to inhibit phosphatase activity. SnRK2s are released to phosphorylate and control the activity of downstream targets to trigger physiological responses. Blue circles indicate ubiquitination, yellow circles indicate phosphorylation events.
When ABA accumulates in cells in response to environmental stress or developmental cues, ABA binds to RCARs and triggers a conformational change that allows the binary complex (receptor–ABA) to physically interact with PP2Cs and inhibit its phosphatase activity. The various RCAR–PP2C heterodimers preclude binding of SnRK2s to PP2Cs, thus stimulating SnRK2s kinase activity. Consequently, SnRK2s are released to phosphorylate and control the activity of downstream factors to trigger physiological responses [19].
3. Role of Ubiquitination and Selective Degradation in ABA Perception and Signaling
Ubiquitination is (next to phosphorylation) another reversible, post-translational modification, originally discovered as a critical element in highly regulated proteolysis and regarded as essential for many other cellular processes. Ubiquitin (Ub) serves as a reusable tag that decides the cellular fate of the proteins and is recognized by numerous Ub-binding proteins directing the target proteins for selective turnover [20][21][22]. Ub, often in the form of polymeric chains, is covalently attached to the target proteins through a three-step E1–E2–E3 conjugation cascade (Figure 2).
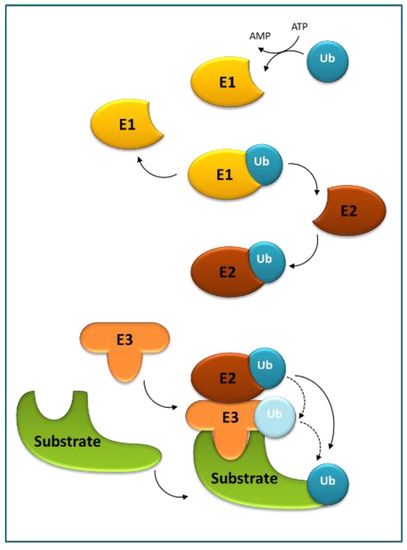
Figure 2. The ubiquitination cascade. Ubiquitin-activating enzyme (E1) activates ubiquitin (Ub) in an ATP-dependent manner, and next the thioester-linked Ub is transferred to Ub-conjugating enzyme (E2). Subsequently, E2 enzymes catalyze the attachment of Ub to a substrate using Ub ligase (E3). E3s, responsible for substrate recognition, transfer Ub either directly from E2 or form an E3–Ub intermediate prior to the transfer.
The process starts with E1 (or Ub-activating enzyme) in which Ub is activated in an ATP-dependent manner, through a thioester bond between the C-terminus of Ub and a cysteine residue of E1, and next the thioester-linked Ub is transferred onto a cysteine residue of E2 (or Ub-conjugating enzyme) by transesterification to form an unstable E2–Ub intermediate. Next, E2 enzymes catalyze the attachment of Ub to lysine (Lys) residues of target proteins using an E3 (or Ub ligase). E3 enzymes are responsible for substrate recognition and either directly transfer the Ub to the substrates from E2 or form an E3–Ub intermediate prior to the transfer. In successive rounds, additional Ub moieties can be attached to one of the seven lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) or the N-terminal methionine (Met1) of Ub to produce poly-Ub chains. The end-product is a Ub–protein conjugate that contains an isopeptide bond between the C-terminal glycine residue of Ub and one of the Lys residues in the substrate. These structurally distinct ubiquitylation patterns are recognized by various effector proteins harboring a ubiquitin-binding domain (UBD) to result in diverse downstream signals. For example, poly-Ub chains are linked by Lys48 target substrates to the 26S proteasome for degradation, whereas poly-Ub chains are linked by Lys63 direct substrates to autophagosomes, both with the concomitant release of Ub moieties for reuse [23].
The E3 Ub ligases can be classified into single- and multi-subunit groups [24]. The single-subunit group includes several subfamilies based upon their mechanisms of action and the presence of specific domains: HECT (homology to E6–AP C terminus), RING (really interesting new gene), and U-box type E3s. The multi-subunit group, cullin-RING box1-ligases (CRLs), are further divided into four subfamilies: SCF (S phase kinase-associated protein 1–cullin 1–F-box), BTB (Bric-a-brac–Tramtrack–Broad complex), DDB (DNA damage-binding domain-containing), and APC (anaphase-promoting complex).
Three E2 enzymes, UBC32, UBC33, and UBC34 play a negative role in drought stress response and ABA signaling [25], and numerous E3 ligases are involved in the inhibition of ABA signaling in optimal conditions, ABA induction upon environmental stress, and ABA attenuation after achieving stress tolerance [8]. The currently known components of ABA perception and signaling and E3 ligases involved in the process are listed in Table 1.
Table 1. ABA pathway components targeted by E3 ligases.
| Pathway Component | Specific Target | E3 Ligase [References] (Remarks) |
|---|---|---|
| ABA Receptors | RCAR1/PYL9 | DDA1 [26]; REA1 [27]; PUB22 [28]; PUB23 [28] |
| RCAR3/PYL8 | RIFP1 [29]; DDA1 [26] | |
| RCAR10/PYL4 | DDA1 [26]; RSL1 [30]; UBC26 (E2)-RFA4 (E3) [31]; RFA1 [31] | |
| RCAR11/PYR1 | RSL1 [30]; RFA1 [31]; RFA4 [31] | |
| Type 2C protein phosphatases (PP2C) | ABI1 | PUB12 [32]; PUB13 [32]; BPM3 [33]; BPM5 [33]; UBC27/AIRP3 [34]; COP1 [35] |
| ABI2 | RGLG1 [36]; RGLG5 [36] | |
| HAB1 | BPM3 [33]; BPM5 [33] | |
| HAB2/NHL29 | RGLG1 [36]; RGLG5 [36] | |
| AHG1 | PIR1.2 [37]; PIR2 [37] | |
| AHG3/PP2CA | RGLG1 [36]; RGLG5 [36]; PIR1.2 [37]; PIR2 [37]; COP1 [35]; BPM3 [33]; BPM5 [33] | |
| HAI1/SAG113 | PIR1.2 [37]; PIR2 [37] | |
| HAI3 | PIR1.2 [37]; PIR2 [37] | |
| SnRK2 kinases | SnRK2.3/SRK2I | PP2-B11 [38] |
| SnRK2.6/OST1 | HOS15 [39] | |
| ABA-responsive transcription factors | ABI5 | DWA1 [40]; DWA2 [40]; KEG [41]; ABD1 [42]; COP9 [43] |
| ABI3 | AIP2 [44][45] | |
| ABI4 | COP1 [46] | |
| ABF1 | KEG [47][48] | |
| ABF2/AREB1 | KEG [49] | |
| ABF3/AREB2 | KEG [47][48] | |
| DREB2 | DRIP1 [50]; DRIP2 [50]; BPM2 [51] | |
| HB6 | BTB1-6/BPM1-6 [52] | |
| MYB96 | MIEL1 [53] (Myb96 activates ABI4) | |
| MYB30 | RHA2b [54]; MIEL1 [53][55] | |
| RAV1 | BPM1 [56][57] | |
| Other regulatory and signaling factors and ABA-responding genes | ADA2B | SKIP24/At1g08710 [58] |
| ATP1/SDIRIP1 | AIRIP2 [59][60]; AIRP1 [59][60]; SDIR1 [61]; (SDIRIP1 activates ABI5) | |
| ACD11 | XBAT35.2 [62]; (ACD11 is one of ABA-responding genes) | |
| RD21 | AIRP3/LOG2 [63]; (positive role in ABA responses; RD21 is Cys protease) | |
| CIPK26 | KEG [64][65] | |
| PP2A | CHIP [66] |
This entry is adapted from the peer-reviewed paper 10.3390/ijms22094638
References
- Orosa, B.; Ustun, S.; Calderon Villalobos, L.I.A.; Genschik, P.; Gibbs, D.; Holdsworth, M.J.; Isono, E.; Lois, M.; Trujillo, M.; Sadanandom, A. Plant proteostasis—Shaping the proteome: A research community aiming to understand molecular mechanisms that control protein abundance. New Phytol. 2020, 227, 1028–1033.
- Bence, N.F.; Sampat, R.M.; Kopito, R.R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 2001, 292, 1552–1555.
- Marshall, R.S.; Vierstra, R.D. Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 2018, 69, 173–208.
- Signorelli, S.; Tarkowski, L.P.; Van den Ende, W.; Bassham, D.C. Linking autophagy to abiotic and biotic stress responses. Trends Plant Sci. 2019, 24, 413–430.
- Li, Y.; Lin, Y.; Li, X.; Guo, S.; Huang, Y.; Xie, Q. Autophagy dances with phytohormones upon multiple stresses. Plants 2020, 9, 1038.
- Finkelstein, R. Abscisic acid synthesis and response. Arab. Book 2013, 11, e0166.
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. Omics approaches toward defining the comprehensive abscisic acid signaling network in plants. Plant Cell Physiol. 2015, 56, 1043–1052.
- Ali, A.; Pardo, J.M.; Yun, D.J. Desensitization of ABA-signaling: The swing from activation to degradation. Front. Plant Sci. 2020, 11, 379.
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54.
- Tal, L.; Gil, M.X.A.; Guercio, A.M.; Shabek, N. Structural aspects of plant hormone signal perception and regulation by ubiquitin ligases. Plant Physiol. 2020, 182, 1537–1544.
- Yu, F.; Xie, Q. Non-26S proteasome endomembrane trafficking pathways in ABA signaling. Trends Plant Sci. 2017, 22, 976–985.
- Yu, F.F.; Xie, Q. Ubiquitination modification precisely modulates the ABA signaling pathway in plants. Yi Chuan 2017, 39, 692–706.
- Bulgakov, V.P.; Avramenko, T.V. Linking brassinosteroid and ABA signaling in the context of stress acclimation. Int. J. Mol. Sci. 2020, 21, 5108.
- Hai, N.N.; Chuong, N.N.; Tu, N.H.C.; Kisiala, A.; Hoang, X.L.T.; Thao, N.P. Role and regulation of cytokinins in plant response to drought stress. Plants 2020, 9, 422.
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86.
- Wang, Q.; Yu, F.; Xie, Q. Balancing growth and adaptation to stress: Crosstalk between brassinosteroid and abscisic acid signaling. Plant Cell Environ. 2020, 43, 2325–2335.
- Fujii, H.; Zhu, J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385.
- Soon, F.F.; Ng, L.M.; Zhou, X.E.; West, G.M.; Kovach, A.; Tan, M.H.; Suino-Powell, K.M.; He, Y.; Xu, Y.; Chalmers, M.J.; et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 2012, 335, 85–88.
- Dejonghe, W.; Okamoto, M.; Cutler, S.R. Small molecule probes of ABA biosynthesis and signaling. Plant Cell Physiol. 2018, 59, 1490–1499.
- Buetow, L.; Huang, D.T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. 2016, 17, 626–642.
- French, M.E.; Koehler, C.F.; Hunter, T. Emerging functions of branched ubiquitin chains. Cell Discov. 2021, 7, 6.
- Zientara-Rytter, K.; Subramani, S. The roles of ubiquitin-binding protein shuttles in the degradative fate of ubiquitinated proteins in the ubiquitin-proteasome system and autophagy. Cells 2019, 8, 40.
- Zientara-Rytter, K.; Sirko, A. To deliver or to degrade—An interplay of the ubiquitin-proteasome system, autophagy and vesicular transport in plants. FEBS J. 2016, 283, 3534–3555.
- Shu, K.; Yang, W. E3 ubiquitin ligases: Ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 2017, 58, 1461–1476.
- Ahn, M.Y.; Oh, T.R.; Seo, D.H.; Kim, J.H.; Cho, N.H.; Kim, W.T. Arabidopsis group XIV ubiquitin-conjugating enzymes AtUBC32, AtUBC33, and AtUBC34 play negative roles in drought stress response. J. Plant Physiol. 2018, 230, 73–79.
- Irigoyen, M.L.; Iniesto, E.; Rodriguez, L.; Puga, M.I.; Yanagawa, Y.; Pick, E.; Strickland, E.; Paz-Ares, J.; Wei, N.; De Jaeger, G.; et al. Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 2014, 26, 712–728.
- Li, D.; Zhang, L.; Li, X.; Kong, X.; Wang, X.; Li, Y.; Liu, Z.; Wang, J.; Li, X.; Yang, Y. AtRAE1 is involved in degradation of ABA receptor RCAR1 and negatively regulates ABA signalling in Arabidopsis. Plant Cell Environ. 2018, 41, 231–244.
- Zhao, J.; Zhao, L.; Zhang, M.; Zafar, S.A.; Fang, J.; Li, M.; Zhang, W.; Li, X. Arabidopsis E3 ubiquitin ligases PUB22 and PUB23 negatively regulate drought tolerance by targeting ABA receptor PYL9 for degradation. Int. J. Mol. Sci. 2017, 18, 1841.
- Li, Y.; Zhang, L.; Li, D.; Liu, Z.; Wang, J.; Li, X.; Yang, Y. The Arabidopsis F-box E3 ligase RIFP1 plays a negative role in abscisic acid signalling by facilitating ABA receptor RCAR3 degradation. Plant Cell Environ. 2016, 39, 571–582.
- Bueso, E.; Rodriguez, L.; Lorenzo-Orts, L.; Gonzalez-Guzman, M.; Sayas, E.; Munoz-Bertomeu, J.; Ibanez, C.; Serrano, R.; Rodriguez, P.L. The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. Cell Mol. Biol. 2014, 80, 1057–1071.
- Fernandez, M.A.; Belda-Palazon, B.; Julian, J.; Coego, A.; Lozano-Juste, J.; Inigo, S.; Rodriguez, L.; Bueso, E.; Goossens, A.; Rodriguez, P.L. RBR-type E3 ligases and the ubiquitin-conjugating enzyme UBC26 regulate abscisic acid receptor levels and signaling. Plant Physiol. 2020, 182, 1723–1742.
- Kong, L.; Cheng, J.; Zhu, Y.; Ding, Y.; Meng, J.; Chen, Z.; Xie, Q.; Guo, Y.; Li, J.; Yang, S.; et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 2015, 6, 8630.
- Julian, J.; Coego, A.; Lozano-Juste, J.; Lechner, E.; Wu, Q.; Zhang, X.; Merilo, E.; Belda-Palazon, B.; Park, S.Y.; Cutler, S.R.; et al. The MATH-BTB BPM3 and BPM5 subunits of Cullin3-RING E3 ubiquitin ligases target PP2CA and other clade A PP2Cs for degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 15725–15734.
- Pan, W.; Lin, B.; Yang, X.; Liu, L.; Xia, R.; Li, J.; Wu, Y.; Xie, Q. The UBC27-AIRP3 ubiquitination complex modulates ABA signaling by promoting the degradation of ABI1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 27694–27702.
- Chen, Q.; Bai, L.; Wang, W.; Shi, H.; Ramon Botella, J.; Zhan, Q.; Liu, K.; Yang, H.Q.; Song, C.P. COP1 promotes ABA-induced stomatal closure by modulating the abundance of ABI/HAB and AHG3 phosphatases. New Phytol. 2021, 229, 2035–2049.
- Wu, Q.; Zhang, X.; Peirats-Llobet, M.; Belda-Palazon, B.; Wang, X.; Cui, S.; Yu, X.; Rodriguez, P.L.; An, C. Ubiquitin ligases RGLG1 and RGLG5 regulate abscisic acid signaling by controlling the turnover of phosphatase PP2CA. Plant Cell 2016, 28, 2178–2196.
- Baek, W.; Lim, C.W.; Luan, S.; Lee, S.C. The RING finger E3 ligases PIR1 and PIR2 mediate PP2CA degradation to enhance abscisic acid response in Arabidopsis. Plant J. Cell Mol. Biol. 2019, 100, 473–486.
- Cheng, C.; Wang, Z.; Ren, Z.; Zhi, L.; Yao, B.; Su, C.; Liu, L.; Li, X. SCFAtPP2-B11 modulates ABA signaling by facilitating SnRK2.3 degradation in Arabidopsis thaliana. PLoS Genet. 2017, 13, e1006947.
- Ali, A.; Kim, J.K.; Jan, M.; Khan, H.A.; Khan, I.U.; Shen, M.; Park, J.; Lim, C.J.; Hussain, S.; Baek, D.; et al. Rheostatic control of ABA signaling through HOS15-mediated OST1 degradation. Mol. Plant 2019, 12, 1447–1462.
- Lee, J.H.; Yoon, H.J.; Terzaghi, W.; Martinez, C.; Dai, M.; Li, J.; Byun, M.O.; Deng, X.W. DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 2010, 22, 1716–1732.
- Stone, S.L.; Williams, L.A.; Farmer, L.M.; Vierstra, R.D.; Callis, J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 2006, 18, 3415–3428.
- Seo, K.I.; Lee, J.H.; Nezames, C.D.; Zhong, S.; Song, E.; Byun, M.O.; Deng, X.W. ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1-based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell 2014, 26, 695–711.
- Jin, D.; Wu, M.; Li, B.; Bucker, B.; Keil, P.; Zhang, S.; Li, J.; Kang, D.; Liu, J.; Dong, J.; et al. The COP9 Signalosome regulates seed germination by facilitating protein degradation of RGL2 and ABI5. PLoS Genet. 2018, 14, e1007237.
- Kurup, S.; Jones, H.D.; Holdsworth, M.J. Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. Cell Mol. Biol. 2000, 21, 143–155.
- Zhang, X.; Garreton, V.; Chua, N.H. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 2005, 19, 1532–1543.
- Xu, X.; Chi, W.; Sun, X.; Feng, P.; Guo, H.; Li, J.; Lin, R.; Lu, C.; Wang, H.; Leister, D.; et al. Convergence of light and chloroplast signals for de-etiolation through ABI4-HY5 and COP1. Nat. Plants 2016, 2, 16066.
- Chen, Y.T.; Liu, H.; Stone, S.; Callis, J. ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis. thaliana. transcription factors ABF1 and ABF3. Plant J. Cell Mol. Biol. 2013, 75, 965–976.
- Liu, H.; Stone, S.L. Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J. Biol. Chem. 2013, 288, 20267–20279.
- Linden, K.J.; Chen, Y.-T.; Kyaw, K.; Schultz, B.; Callis, J. Factors that affect protein abundance of the bZIP transcription factor ABRE-BINDING FACTOR 2 (ABF2), a positive regulator of abscisic acid signaling. bioRxiv 2021.
- Qin, F.; Sakuma, Y.; Tran, L.S.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Fujita, M.; Umezawa, T.; Sawano, Y.; Miyazono, K.; et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 2008, 20, 1693–1707.
- Morimoto, K.; Ohama, N.; Kidokoro, S.; Mizoi, J.; Takahashi, F.; Todaka, D.; Mogami, J.; Sato, H.; Qin, F.; Kim, J.S.; et al. BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E8528–E8536.
- Lechner, E.; Leonhardt, N.; Eisler, H.; Parmentier, Y.; Alioua, M.; Jacquet, H.; Leung, J.; Genschik, P. MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev. Cell 2011, 21, 1116–1128.
- Lee, H.G.; Seo, P.J. The Arabidopsis MIEL1 E3 ligase negatively regulates ABA signalling by promoting protein turnover of MYB96. Nat. Commun. 2016, 7, 12525.
- Zheng, Y.; Chen, Z.; Ma, L.; Liao, C. The ubiquitin E3 ligase RHA2b promotes degradation of MYB30 in abscisic acid signaling. Plant Physiol. 2018, 178, 428–440.
- Marino, D.; Froidure, S.; Canonne, J.; Ben Khaled, S.; Khafif, M.; Pouzet, C.; Jauneau, A.; Roby, D.; Rivas, S. Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat. Commun. 2013, 4, 1476.
- Chen, L.; Lee, J.H.; Weber, H.; Tohge, T.; Witt, S.; Roje, S.; Fernie, A.R.; Hellmann, H. Arabidopsis BPM proteins function as substrate adaptors to a cullin3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell 2013, 25, 2253–2264.
- Feng, C.Z.; Chen, Y.; Wang, C.; Kong, Y.H.; Wu, W.H.; Chen, Y.F. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. Cell Mol. Biol. 2014, 80, 654–668.
- Rao, V.; Virupapuram, V. Arabidopsis F-box protein At1g08710 interacts with transcriptional protein ADA2b and imparts drought stress tolerance by negatively regulating seedling growth. Biochem. Biophys. Res. Commun. 2021, 536, 45–51.
- Cho, S.K.; Ryu, M.Y.; Seo, D.H.; Kang, B.G.; Kim, W.T. The Arabidopsis RING E3 ubiquitin ligase AtAIRP2 plays combinatory roles with AtAIRP1 in abscisic acid-mediated drought stress responses. Plant Physiol. 2011, 157, 2240–2257.
- Oh, T.R.; Kim, J.H.; Cho, S.K.; Ryu, M.Y.; Yang, S.W.; Kim, W.T. AtAIRP2 E3 ligase affects ABA and high-salinity responses by stimulating its ATP1/SDIRIP1 substrate turnover. Plant Physiol. 2017, 174, 2515–2531.
- Zhang, H.; Cui, F.; Wu, Y.; Lou, L.; Liu, L.; Tian, M.; Ning, Y.; Shu, K.; Tang, S.; Xie, Q. The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 2015, 27, 214–227.
- Li, Q.; Serio, R.J.; Schofield, A.; Liu, H.; Rasmussen, S.R.; Hofius, D.; Stone, S.L. Arabidopsis RING-type E3 ubiquitin ligase XBAT35.2 promotes proteasome-dependent degradation of ACD11 to attenuate abiotic stress tolerance. Plant J. Cell Mol. Biol. 2020, 104, 1712–1723.
- Kim, J.H.; Kim, W.T. The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high-salt and drought stress responses. Plant Physiol. 2013, 162, 1733–1749.
- Lyzenga, W.J.; Liu, H.; Schofield, A.; Muise-Hennessey, A.; Stone, S.L. Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin-proteasome system. J. Exp. Bot. 2013, 64, 2779–2791.
- Lyzenga, W.J.; Sullivan, V.; Liu, H.; Stone, S.L. The kinase activity of Calcineurin B-like Interacting Protein Kinase 26 (CIPK26) influences its own stability and that of the ABA-regulated ubiquitin ligase, Keep on Going (KEG). Front. Plant Sci. 2017, 8, 502.
- Luo, J.; Shen, G.; Yan, J.; He, C.; Zhang, H. AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J. Cell Mol. Biol. 2006, 46, 649–657.
This entry is offline, you can click here to edit this entry!
