Nanobodies are highly water-soluble and stable, have high specificity, and can bind their targets with very high affinity, often in the low nanomolar range.
- nanobodies
- VHHs
- molecular imaging
- radionuclide imaging
- immuno-PET
- SPECT/CT
- cancer-specific markers
- immune checkpoint imaging
1. Introduction
In addition to conventional antibodies, camelids, such as llamas and alpacas, have unique heavy-chain-only antibodies [1]. These antibodies are unique in that the variable regions are encompassed by a single domain (VHH) instead of two separate domains (VH and VL) as seen in conventional antibodies [2]. The variable domains of the camelid heavy-chain-only antibodies have found widespread applications in biomedical research.
Nanobodies are highly water-soluble and stable, have high specificity, and can bind to their targets with high affinity, often in the low nanomolar range [3]. VHHs are stable as single-domain antibodies because of several mutations on their surface that allow them to be water soluble [3]. In particular, several residues that would be at the VH–VL interface in conventional antibodies are mutated for hydrophobic to hydrophilic residues (G44E, L45R, and W47G) (Figure 2), enhancing their stability and solubility as a single domain. In addition, there is a solubility enhancing mutation, most commonly found in camel VHHs, at the VH–CH1 interface (L11S) (Figure 2A,C).
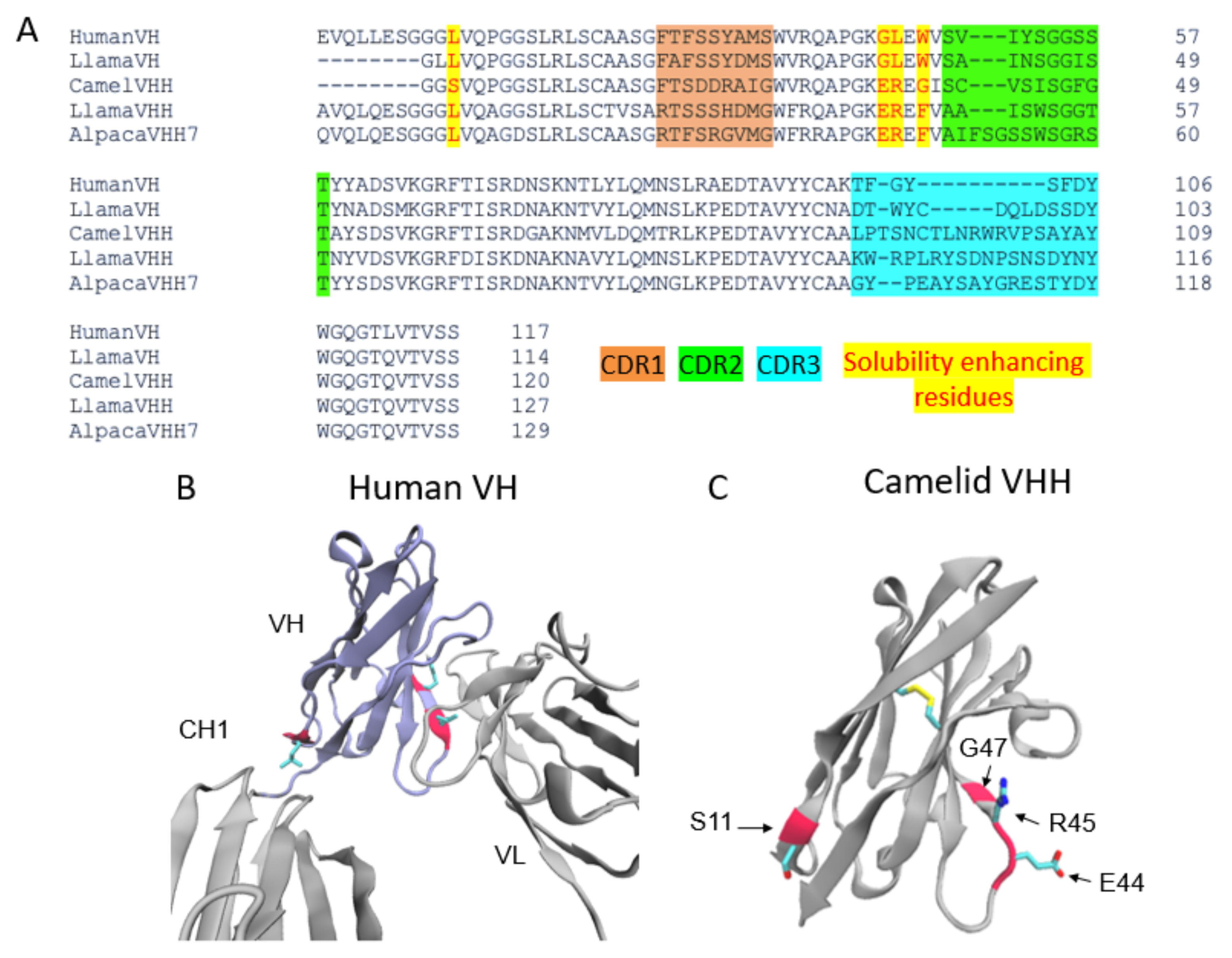
The factor contributing to the high affinity of these nanobodies is that their frameworks have three complementarity-determining regions (CDRs). These CDRs are analogous to those found in human antibody VH and VL domains and are subject to somatic hypermutation in the course of affinity maturation. The CDR3 of VHHs is especially long in comparison to the human counterpart [4]. The length and flexibility of VHH CDR3s enable the nanobody to access a variety of conformations. In some cases, VHH CDR3s are able to fold back and make contact with the nanobody framework [4]. Taken together, these factors compensate for the lack of sequence variability incurred by the loss of VL CDRs, allowing VHHs to bind to their targets with high specificity and affinity (Figure 1C,D and Figure 2A).
Methods of generating nanobodies against an antigen of interest have already been well established [2]. In brief, a llama or alpaca (among a variety of other camelids) is immunized against the antigen(s) of interest [2]. Administration of the protein antigen is typically accompanied by an immune adjuvant that serves to enhance the overall immune response [5]. Several weeks later, blood is harvested from the immunized animal and peripheral blood mononuclear cells (PBMCs) are purified. This purification is then followed by total RNA extraction, VHH amplification, and finally, the construction of a phage display library. Phage display libraries are among the most common methods of preparing nanobody libraries, but other methods, such as E. coli or yeast display, could alternatively be used [2][6][7]. Finally, the lead VHHs are identified and expressed as soluble proteins using reliable approaches, such as magnetic-activated cell sorting (MACS), fluorescence-activated cell sorting (FACS), or panning against immobilized antigens (Figure 3) [8][9][10].
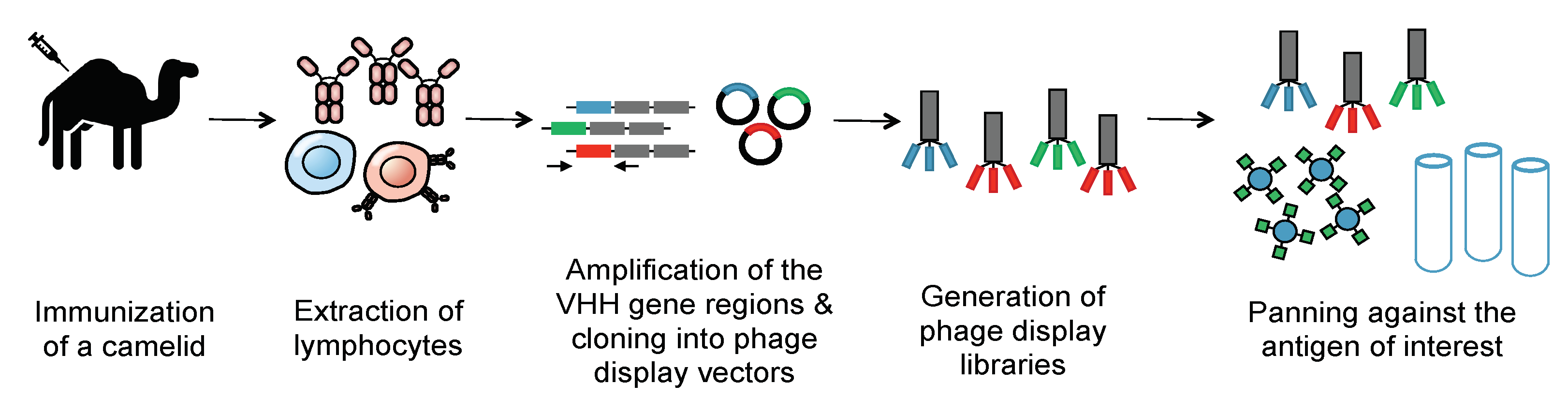
Figure 3. Generation of a nanobody library. To create an immune library, camelids are immunized against a molecule of interest. mRNA of the camelids’ peripheral blood mononuclear cells is then converted into cDNA. PCR is then employed to amplify the VHH genes. These immune VHH genes will then be cloned into a phage display vector. Phages are then generated using E. coli strains such as TG1. Phage libraries are then panned against immobilized antigens to select for nanobodies that selectively bind the antigen with high affinity. The panned libraries are then used for reinfection of E. coli to obtain specific clones.
The short circulatory half-life of nanobodies have allowed the use of a range of isotopes with short half-lives for imaging, such as Galium-68 (68Ga, t1/2 = 67.71 min) and 18F (t1/2 = 109.7 min), as well as longer-lived isotopes, such as Technetium-99m (99mTc t1/2 = 6.0 h), Copper-64 (64Cu t1/2 = 12.7 h), Indium-111 (111In t1/2 = 67.2 h), Zirconium-89 (89Zr t1/2 = 78.41 h), and Lutetium-177 (177Lu t1/2 = 6.7 days). Similar to other antibody fragments, nanobodies are commonly labeled nonspecifically via their side-chain lysine residues using chelators or radioisotopes that are functionalized with amine-reactive groups such as N-hydroxysuccinimide (NHS) or isothiocyanatobenzyl (pSCN) groups. While this strategy is robust and reproducible, it is not site-specific, which may damage antigen-binding sites [11]. To address this issue and to ensure the binding capacity is not compromised, a variety of site-specific labeling approaches, such as the use of sortase technology, have been developed [12]. Another common approach is using a His6 tag to install 99mTc, a commonly used SPECT isotope [13].
2. Nanobodies for Medical Imaging
Biopsies will likely remain the gold standard of cancer diagnostics for the foreseeable future; however, biopsies can sometimes be unrepresentative of the greater TME or targeted organ. Non-invasive immuno-PET imaging, as an adjunct to biopsies, can provide a holistic view of the TME and offer a complete insight into both primary and metastatic tumors. Information revealed via imaging can help to make informed treatment decisions. Imaging is also beneficial in understanding the progression and pathogenesis of a variety of diseases, such as fibrosis, cardiovascular complications, arthritis, and neurological diseases. Therefore, immuno-PET imaging is a potentially revolutionary addition to disease management and treatment.
| Target | Agent | Reactivity | Clinical Trials: Stage and Status (If Applicable) | References |
|---|---|---|---|---|
| EGFR | 99mTc-8B6 | Human | Preclinical | [7] |
| 99mTc-7C12 | Human | Preclinical | [21] | |
| HER2 | 177Lu-2Rs15dHIS | Human | Preclinical | [22] |
| 18F-FB-2Rs15d | Murine | Preclinical | [23] | |
| 18F-RL-I-5F7 | Murine | Preclinical | [24] | |
| 68Ga-2Rs15d | Human | Clinical | [23][25] | |
| HER3 | 89Zr-MSB0010853 | Murine | Preclinical | [26] |
| CEA | 99mTc-NbCEA5 | Human | Preclinical | [27] |
| PSMA | 111In-JVZ007 | Human | Preclinical | [28] |
| HGF | 89Zr-1E2, 89Zr-6E10 | Human | Preclinical | [29] |
| CD20 | 68Ga-9079 | Human | Preclinical | [30] |
| CD38 | 68Ga-NOTA-Nb1053 | Murine | Preclinical | [31] |
| Mesothelin | 99mTc-A1, 99mTc-C6 | Human | Preclinical | [32] |
| MMR | 99mTc-d a-MMR Nb cl1 | Murine | Preclinical | [33][34] |
| 18 F-FB-anti-MMR 3.49 | Human, Murine | Preclinical | [35] | |
| 68Ga-NOTA-Anti-MMR-VHH2 | Human | Clinical, NCT04168528 (Active) | [36] | |
| MHC II | [18F]FDG -VHH7 | Murine | Preclinical | [37] |
| 64Cu- VHH4 | Human | Preclinical | [38] | |
| CD11b | 89Zr-VHHDC13 (PEGylated) |
Murine | Preclinical | [39] |
| 18F-VHHDC13 | Human | Preclinical | [40] | |
| CD8 | 89Zr-VHH-X118 (PEGylated) |
Murine | Preclinical | [12] |
| 68Ga-NOTA-SNA006 | Human | Preclinical | [41] | |
| Mouse Dendritic Cells | 99mTc-Nb-DC2.1 | Murine | Preclinical | [42] |
| 99mTc-Nb-DC1.8 | Murine | Preclinical | [42] | |
| PD-L1 | 18F-B3, 18F-A12, 64Cu-B3 | Murine | Preclinical | [43] |
| 99mTc-C3, 99mTc-C7, 99mTc-E2, 99mTc-E4, 99mTc-K2 | Murine | Preclinical | [44][45][46][47] | |
| 68Ga-NOTA-Nb109 | Human | Preclinical | [48] | |
| 99mTc-NM-01 | Human | Clinical, NCT02978196 (Concluded) |
[49] | |
| 89Zr-envafolimab (Fc fusion) |
Human | Clinical, NCT03638804 (Active) |
[50][51] | |
| CTLA-4 | 18F-H11, 89Zr-H11 | Murine | Preclinical | [51][52] |
| LAG-3 | 99mTc-anti-moLAG-3 3206, 99mTc-anti-moLAG-3 3208, 99mTc-anti-moLAG-3 3132, 99mTc-anti-moLAG-3 3141 |
Murine | Preclinical | [53][54] |
| VCAM-1 | 99m Tc-cAbVCAM1-5 | Human, Murine | Preclinical | [33][55][56][57] |
| FN-EIIIB (ECM) | 64 Cu-NJB2 | Human, Murine | Preclinical | [58] |
| αSyn | NbSyn2, NbSyn87 (fused to fluorescent proteins for imaging) | Human | Preclinical | [59][60] |
| DPP6 | 99m Tc-4hD29 | Human | Preclinical | [61] |
| Vsig4 | 99m Tc-NbV4 | Murine | Preclinical | [62][63] |
| Clec4F (KC) | 99m Tc-NbC4 | Murine | Preclinical | [62] |
This entry is adapted from the peer-reviewed paper 10.3390/biom11050637
References
- Bannas, P.; Hambach, J.; Koch-Nolte, F. Nanobodies and Nanobody-Based Human Heavy Chain Antibodies As Antitumor Therapeutics. Front. Immunol. 2017, 8, 1603.
- Pardon, E.; Laeremans, T.; Triest, S.; Rasmussen, S.G.F.; Wohlkönig, A.; Ruf, A.; Muyldermans, S.; Hol, W.G.J.; Kobilka, B.K.; Steyaert, J. A General Protocol for the Generation of Nanobodies for Structural Biology. Nat. Protoc. 2014, 9, 674–693.
- Debie, P.; Devoogdt, N.; Hernot, S. Targeted Nanobody-Based Molecular Tracers for Nuclear Imaging and Image-Guided Surgery. Antibodies 2019, 8, 12.
- Mitchell, L.S.; Colwell, L.J. Comparative Analysis of Nanobody Sequence and Structure Data. Proteins Struct. Funct. Bioinform. 2018, 86, 697–706.
- Muyldermans, S. A Guide to: Generation and Design of Nanobodies. FEBS J. 2020.
- Fleetwood, F.; Devoogdt, N.; Pellis, M.; Wernery, U.; Muyldermans, S.; Ståhl, S.; Löfblom, J. Surface Display of a Single-Domain Antibody Library on Gram-Positive Bacteria. Cell. Mol. Life Sci. CMLS 2013, 70, 1081–1093.
- Koide, A.; Koide, S. Affinity Maturation of Single-Domain Antibodies by Yeast Surface Display. Methods Mol. Biol. Clifton NJ 2012, 911, 431–443.
- Salema, V.; Fernández, L.Á. Escherichia Coli Surface Display for the Selection of Nanobodies. Microb. Biotechnol. 2017, 10, 1468–1484.
- Salema, V.; Mañas, C.; Cerdán, L.; Piñero-Lambea, C.; Marín, E.; Roovers, R.C.; Van Bergen En Henegouwen, P.M.P.; Fernández, L.Á. High Affinity Nanobodies against Human Epidermal Growth Factor Receptor Selected on Cells by E. coli Display. mAbs 2016, 8, 1286–1301.
- McMahon, C.; Baier, A.S.; Pascolutti, R.; Wegrecki, M.; Zheng, S.; Ong, J.X.; Erlandson, S.C.; Hilger, D.; Rasmussen, S.G.F.; Ring, A.M.; et al. Yeast Surface Display Platform for Rapid Discovery of Conformationally Selective Nanobodies. Nat. Struct. Mol. Biol. 2018, 25, 289–296.
- Higashikawa, K.; Yagi, K.; Watanabe, K.; Kamino, S.; Ueda, M.; Hiromura, M.; Enomoto, S. 64Cu-DOTA-Anti-CTLA-4 MAb Enabled PET Visualization of CTLA-4 on the T-Cell Infiltrating Tumor Tissues. PLoS ONE 2014, 9, e109866.
- Rashidian, M.; Ingram, J.R.; Dougan, M.; Dongre, A.; Whang, K.A.; LeGall, C.; Cragnolini, J.J.; Bierie, B.; Gostissa, M.; Gorman, J.; et al. Predicting the Response to CTLA-4 Blockade by Longitudinal Noninvasive Monitoring of CD8 T Cells. J. Exp. Med. 2017, 214, 2243–2255.
- Xavier, C.; Devoogdt, N.; Hernot, S.; Vaneycken, I.; D’Huyvetter, M.; De Vos, J.; Massa, S.; Lahoutte, T.; Caveliers, V. Site-Specific Labeling of His-Tagged Nanobodies with 99mTc: A Practical Guide. Methods Mol. Biol. Clifton NJ 2012, 911, 485–490.
- Lambert, B.; Cybulla, M.; Weiner, S.M.; Van De Wiele, C.; Ham, H.; Dierckx, R.A.; Otte, A. Renal Toxicity after Radionuclide Therapy. Radiat. Res. 2004, 161, 607–611.
- Vegt, E.; de Jong, M.; Wetzels, J.F.M.; Masereeuw, R.; Melis, M.; Oyen, W.J.G.; Gotthardt, M.; Boerman, O.C. Renal Toxicity of Radiolabeled Peptides and Antibody Fragments: Mechanisms, Impact on Radionuclide Therapy, and Strategies for Prevention. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2010, 51, 1049–1058.
- van Eerd, J.E.M.; Vegt, E.; Wetzels, J.F.M.; Russel, F.G.M.; Masereeuw, R.; Corstens, F.H.M.; Oyen, W.J.G.; Boerman, O.C. Gelatin-Based Plasma Expander Effectively Reduces Renal Uptake of 111In-Octreotide in Mice and Rats. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2006, 47, 528–533.
- Vegt, E.; Wetzels, J.F.M.; Russel, F.G.M.; Masereeuw, R.; Boerman, O.C.; van Eerd, J.E.; Corstens, F.H.M.; Oyen, W.J.G. Renal Uptake of Radiolabeled Octreotide in Human Subjects Is Efficiently Inhibited by Succinylated Gelatin. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2006, 47, 432–436.
- Jovčevska, I.; Muyldermans, S. The Therapeutic Potential of Nanobodies. BioDrugs 2020, 34, 11–26.
- Ackaert, C.; Smiejkowska, N.; Xavier, C.; Sterckx, Y.G.J.; Denies, S.; Stijlemans, B.; Elkrim, Y.; Devoogdt, N.; Caveliers, V.; Lahoutte, T.; et al. Immunogenicity Risk Profile of Nanobodies. Front. Immunol. 2021, 12, 632687.
- Zimmermann, I.; Egloff, P.; Hutter, C.A.J.; Kuhn, B.T.; Bräuer, P.; Newstead, S.; Dawson, R.J.P.; Geertsma, E.R.; Seeger, M.A. Generation of Synthetic Nanobodies against Delicate Proteins. Nat. Protoc. 2020, 15, 1707–1741.
- Gainkam, L.O.T.; Keyaerts, M.; Caveliers, V.; Devoogdt, N.; Vanhove, C.; Van Grunsven, L.; Muyldermans, S.; Lahoutte, T. Correlation between Epidermal Growth Factor Receptor-Specific Nanobody Uptake and Tumor Burden: A Tool for Noninvasive Monitoring of Tumor Response to Therapy. Mol. Imaging Biol. 2011, 13, 940–948.
- D’Huyvetter, M.; Aerts, A.; Xavier, C.; Vaneycken, I.; Devoogdt, N.; Gijs, M.; Impens, N.; Baatout, S.; Ponsard, B.; Muyldermans, S.; et al. Development of 177Lu-Nanobodies for Radioimmunotherapy of HER2-Positive Breast Cancer: Evaluation of Different Bifunctional Chelators: 177LU-NANOBODIES FOR RADIOIMMUNOTHERAPY. Contrast Media Mol. Imaging 2012, 7, 254–264.
- Xavier, C.; Blykers, A.; Vaneycken, I.; D’Huyvetter, M.; Heemskerk, J.; Lahoutte, T.; Devoogdt, N.; Caveliers, V. (18)F-Nanobody for PET Imaging of HER2 Overexpressing Tumors. Nucl. Med. Biol. 2016, 43, 247–252.
- Vaidyanathan, G.; McDougald, D.; Choi, J.; Koumarianou, E.; Weitzel, D.; Osada, T.; Lyerly, H.K.; Zalutsky, M.R. Preclinical Evaluation of 18F-Labeled Anti-HER2 Nanobody Conjugates for Imaging HER2 Receptor Expression by Immuno-PET. J. Nucl. Med. 2016, 57, 967–973.
- Zhou, Z.; Vaidyanathan, G.; McDougald, D.; Kang, C.M.; Balyasnikova, I.; Devoogdt, N.; Ta, A.N.; McNaughton, B.R.; Zalutsky, M.R. Fluorine-18 Labeling of the HER2-Targeting Single-Domain Antibody 2Rs15d Using a Residualizing Label and Preclinical Evaluation. Mol. Imaging Biol. 2017, 19, 867–877.
- Warnders, F.J.; Terwisscha van Scheltinga, A.G.T.; Knuehl, C.; van Roy, M.; de Vries, E.F.J.; Kosterink, J.G.W.; de Vries, E.G.E.; Lub-de Hooge, M.N. Human Epidermal Growth Factor Receptor 3-Specific Tumor Uptake and Biodistribution of 89Zr-MSB0010853 Visualized by Real-Time and Noninvasive PET Imaging. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2017, 58, 1210–1215.
- Vaneycken, I.; Govaert, J.; Vincke, C.; Caveliers, V.; Lahoutte, T.; De Baetselier, P.; Raes, G.; Bossuyt, A.; Muyldermans, S.; Devoogdt, N. In Vitro Analysis and in Vivo Tumor Targeting of a Humanized, Grafted Nanobody in Mice Using Pinhole SPECT/Micro-CT. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2010, 51, 1099–1106.
- Chatalic, K.L.S.; Veldhoven-Zweistra, J.; Bolkestein, M.; Hoeben, S.; Koning, G.A.; Boerman, O.C.; de Jong, M.; van Weerden, W.M. A Novel 111In-Labeled Anti-Prostate-Specific Membrane Antigen Nanobody for Targeted SPECT/CT Imaging of Prostate Cancer. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2015, 56, 1094–1099.
- Vosjan, M.J.W.D.; Vercammen, J.; Kolkman, J.A.; Stigter-van Walsum, M.; Revets, H.; van Dongen, G.A.M.S. Nanobodies Targeting the Hepatocyte Growth Factor: Potential New Drugs for Molecular Cancer Therapy. Mol. Cancer Ther. 2012, 11, 1017–1025.
- Krasniqi, A.; D’Huyvetter, M.; Xavier, C.; Van der Jeught, K.; Muyldermans, S.; Van Der Heyden, J.; Lahoutte, T.; Tavernier, J.; Devoogdt, N. Theranostic Radiolabeled Anti-CD20 SdAb for Targeted Radionuclide Therapy of Non-Hodgkin Lymphoma. Mol. Cancer Ther. 2017, 16, 2828–2839.
- Wang, C.; Chen, Y.; Hou, Y.N.; Liu, Q.; Zhang, D.; Zhao, H.; Zhang, Y.; An, S.; Li, L.; Hou, J.; et al. ImmunoPET Imaging of Multiple Myeloma with [68Ga]Ga-NOTA-Nb1053. Eur. J. Nucl. Med. Mol. Imaging 2021.
- Montemagno, C.; Bacot, S.; Ahmadi, M.; Kerfelec, B.; Baty, D.; Debiossat, M.; Soubies, A.; Perret, P.; Riou, L.; Fagret, D.; et al. Preclinical Evaluation of Mesothelin-Specific Ligands for SPECT Imaging of Triple-Negative Breast Cancer. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2018, 59, 1056–1062.
- Put, S.; Schoonooghe, S.; Devoogdt, N.; Schurgers, E.; Avau, A.; Mitera, T.; D’Huyvetter, M.; De Baetselier, P.; Raes, G.; Lahoutte, T.; et al. SPECT Imaging of Joint Inflammation with Nanobodies Targeting the Macrophage Mannose Receptor in a Mouse Model for Rheumatoid Arthritis. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2013, 54, 807–814.
- Movahedi, K.; Schoonooghe, S.; Laoui, D.; Houbracken, I.; Waelput, W.; Breckpot, K.; Bouwens, L.; Lahoutte, T.; Baetselier, P.D.; Raes, G.; et al. Nanobody-Based Targeting of the Macrophage Mannose Receptor for Effective In Vivo Imaging of Tumor-Associated Macrophages. Cancer Res. 2012, 72, 4165–4177.
- Blykers, A.; Schoonooghe, S.; Xavier, C.; D’hoe, K.; Laoui, D.; D’Huyvetter, M.; Vaneycken, I.; Cleeren, F.; Bormans, G.; Heemskerk, J.; et al. PET Imaging of Macrophage Mannose Receptor-Expressing Macrophages in Tumor Stroma Using 18F-Radiolabeled Camelid Single-Domain Antibody Fragments. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2015, 56, 1265–1271.
- Xavier, C.; Blykers, A.; Laoui, D.; Bolli, E.; Vaneyken, I.; Bridoux, J.; Baudhuin, H.; Raes, G.; Everaert, H.; Movahedi, K.; et al. Clinical Translation of [68Ga]Ga-NOTA-Anti-MMR-SdAb for PET/CT Imaging of Protumorigenic Macrophages. Mol. Imaging Biol. 2019, 21, 898–906.
- Rashidian, M.; Keliher, E.; Dougan, M.; Juras, P.K.; Cavallari, M.; Wojtkiewicz, G.R.; Jacobsen, J.; Edens, J.G.; Tas, J.M.G.; Victora, G.; et al. The Use of (18)F-2-Fluorodeoxyglucose (FDG) to Label Antibody Fragments for Immuno-PET of Pancreatic Cancer. ACS Cent. Sci. 2015, 1, 142–147.
- Van Elssen, C.H.M.J.; Rashidian, M.; Vrbanac, V.; Wucherpfennig, K.W.; Habre, Z.E.; Sticht, J.; Freund, C.; Jacobsen, J.T.; Cragnolini, J.; Ingram, J.; et al. Noninvasive Imaging of Human Immune Responses in a Human Xenograft Model of Graft-Versus-Host Disease. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2017, 58, 1003–1008.
- Rashidian, M.; LaFleur, M.W.; Verschoor, V.L.; Dongre, A.; Zhang, Y.; Nguyen, T.H.; Kolifrath, S.; Aref, A.R.; Lau, C.J.; Paweletz, C.P.; et al. Immuno-PET Identifies the Myeloid Compartment as a Key Contributor to the Outcome of the Antitumor Response under PD-1 Blockade. Proc. Natl. Acad. Sci. USA 2019, 116, 16971–16980.
- Rashidian, M.; Keliher, E.J.; Bilate, A.M.; Duarte, J.N.; Wojtkiewicz, G.R.; Jacobsen, J.T.; Cragnolini, J.; Swee, L.K.; Victora, G.D.; Weissleder, R.; et al. Noninvasive Imaging of Immune Responses. Proc. Natl. Acad. Sci. USA 2015, 112, 6146–6151.
- Zhao, H.; Wang, C.; Yang, Y.; Sun, Y.; Wei, W.; Wang, C.; Wan, L.; Zhu, C.; Li, L.; Huang, G.; et al. ImmunoPET Imaging of Human CD8+ T Cells with Novel 68Ga-Labeled Nanobody Companion Diagnostic Agents. J. Nanobiotechnology 2021, 19, 42.
- Groeve, K.D.; Deschacht, N.; Koninck, C.D.; Caveliers, V.; Lahoutte, T.; Devoogdt, N.; Muyldermans, S.; Baetselier, P.D.; Raes, G. Nanobodies as Tools for In Vivo Imaging of Specific Immune Cell Types. J. Nucl. Med. 2010, 51, 782–789.
- Ingram, J.R.; Dougan, M.; Rashidian, M.; Knoll, M.; Keliher, E.J.; Garrett, S.; Garforth, S.; Blomberg, O.S.; Espinosa, C.; Bhan, A.; et al. PD-L1 Is an Activation-Independent Marker of Brown Adipocytes. Nat. Commun. 2017, 8, 647.
- Broos, K.; Keyaerts, M.; Lecocq, Q.; Renmans, D.; Nguyen, T.; Escors, D.; Liston, A.; Raes, G.; Breckpot, K.; Devoogdt, N. Non-Invasive Assessment of Murine PD-L1 Levels in Syngeneic Tumor Models by Nuclear Imaging with Nanobody Tracers. Oncotarget 2017, 8, 41932–41946.
- Broos, K.; Lecocq, Q.; Raes, G.; Devoogdt, N.; Keyaerts, M.; Breckpot, K. Noninvasive Imaging of the PD-1:PD-L1 Immune Checkpoint: Embracing Nuclear Medicine for the Benefit of Personalized Immunotherapy. Theranostics 2018, 8, 3559–3570.
- Bridoux, J.; Broos, K.; Lecocq, Q.; Debie, P.; Martin, C.; Ballet, S.; Raes, G.; Neyt, S.; Vanhove, C.; Breckpot, K.; et al. Anti-Human PD-L1 Nanobody for Immuno-PET Imaging: Validation of a Conjugation Strategy for Clinical Translation. Biomolecules 2020, 10, 1388.
- Broos, K.; Lecocq, Q.; Xavier, C.; Bridoux, J.; Nguyen, T.T.; Corthals, J.; Schoonooghe, S.; Lion, E.; Raes, G.; Keyaerts, M.; et al. Evaluating a Single Domain Antibody Targeting Human PD-L1 as a Nuclear Imaging and Therapeutic Agent. Cancers 2019, 11, 872.
- Liu, Q.; Jiang, L.; Li, K.; Li, H.; Lv, G.; Lin, J.; Qiu, L. Immuno-PET Imaging of 68Ga-Labeled Nanobody Nb109 for Dynamic Monitoring the PD-L1 Expression in Cancers. Cancer Immunol. Immunother. CII 2021.
- Xing, Y.; Chand, G.; Liu, C.; Cook, G.J.R.; O’Doherty, J.; Zhao, L.; Wong, N.C.L.; Meszaros, L.K.; Ting, H.H.; Zhao, J. Early Phase I Study of a 99mTc-Labeled Anti–Programmed Death Ligand-1 (PD-L1) Single-Domain Antibody in SPECT/CT Assessment of PD-L1 Expression in Non–Small Cell Lung Cancer. J. Nucl. Med. 2019, 60, 1213–1220.
- Li, D.; Zou, S.; Cheng, S.; Song, S.; Wang, P.; Zhu, X. Monitoring the Response of PD-L1 Expression to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Nonsmall-Cell Lung Cancer Xenografts by Immuno-PET Imaging. Mol. Pharm. 2019, 16, 3469–3476.
- Li, D.; Cheng, S.; Zou, S.; Zhu, D.; Zhu, T.; Wang, P.; Zhu, X. Immuno-PET Imaging of 89Zr Labeled Anti-PD-L1 Domain Antibody. Mol. Pharm. 2018, 15, 1674–1681.
- Ingram, J.R.; Blomberg, O.S.; Rashidian, M.; Ali, L.; Garforth, S.; Fedorov, E.; Fedorov, A.A.; Bonanno, J.B.; Le Gall, C.; Crowley, S.; et al. Anti-CTLA-4 Therapy Requires an Fc Domain for Efficacy. Proc. Natl. Acad. Sci. USA 2018, 115, 3912–3917.
- Lecocq, Q.; Zeven, K.; De Vlaeminck, Y.; Martens, S.; Massa, S.; Goyvaerts, C.; Raes, G.; Keyaerts, M.; Breckpot, K.; Devoogdt, N. Noninvasive Imaging of the Immune Checkpoint LAG-3 Using Nanobodies, from Development to Pre-Clinical Use. Biomolecules 2019, 9, 548.
- Lecocq, Q.; Awad, R.M.; De Vlaeminck, Y.; De Mey, W.; Ertveldt, T.; Goyvaerts, C.; Raes, G.; Thielemans, K.; Keyaerts, M.; Devoogdt, N.; et al. Nanobody Nuclear Imaging Allows Noninvasive Quantification of LAG-3 Expression by Tumor-Infiltrating Leukocytes and Predicts Response of Immune Checkpoint Blockade. J. Nucl. Med. 2021.
- Broisat, A.; Hernot, S.; Toczek, J.; De Vos, J.; Riou, L.M.; Martin, S.; Ahmadi, M.; Thielens, N.; Wernery, U.; Caveliers, V.; et al. Nanobodies Targeting Mouse/Human VCAM1 for the Nuclear Imaging of Atherosclerotic Lesions. Circ. Res. 2012, 110, 927–937.
- Sun Yoo, J.; Lee, J.; Ho Jung, J.; Seok Moon, B.; Kim, S.; Chul Lee, B.; Eun Kim, S. SPECT/CT Imaging of High-Risk Atherosclerotic Plaques Using Integrin-Binding RGD Dimer Peptides. Sci. Rep. 2015, 5, 11752.
- Chakravarty, R.; Goel, S.; Cai, W. Nanobody: The “Magic Bullet” for Molecular Imaging? Theranostics 2014, 4, 386–398.
- Jailkhani, N.; Ingram, J.R.; Rashidian, M.; Rickelt, S.; Tian, C.; Mak, H.; Jiang, Z.; Ploegh, H.L.; Hynes, R.O. Noninvasive Imaging of Tumor Progression, Metastasis, and Fibrosis Using a Nanobody Targeting the Extracellular Matrix. Proc. Natl. Acad. Sci. USA 2019, 116, 14181–14190.
- Gerdes, C.; Waal, N.; Offner, T.; Fornasiero, E.F.; Wender, N.; Verbarg, H.; Manzini, I.; Trenkwalder, C.; Mollenhauer, B.; Strohäker, T.; et al. A Nanobody-Based Fluorescent Reporter Reveals Human α-Synuclein in the Cell Cytosol. Nat. Commun. 2020, 11, 2729.
- Iljina, M.; Hong, L.; Horrocks, M.H.; Ludtmann, M.H.; Choi, M.L.; Hughes, C.D.; Ruggeri, F.S.; Guilliams, T.; Buell, A.K.; Lee, J.-E.; et al. Nanobodies Raised against Monomeric Ɑ-Synuclein Inhibit Fibril Formation and Destabilize Toxic Oligomeric Species. BMC Biol. 2017, 15, 57.
- Balhuizen, A.; Massa, S.; Mathijs, I.; Turatsinze, J.-V.; De Vos, J.; Demine, S.; Xavier, C.; Villate, O.; Millard, I.; Egrise, D.; et al. A Nanobody-Based Tracer Targeting DPP6 for Non-Invasive Imaging of Human Pancreatic Endocrine Cells. Sci. Rep. 2017, 7, 15130.
- Zheng, F.; Sparkes, A.; De Baetselier, P.; Schoonooghe, S.; Stijlemans, B.; Muyldermans, S.; Flamand, V.; Van Ginderachter, J.A.; Devoogdt, N.; Raes, G.; et al. Molecular Imaging with Kupffer Cell-Targeting Nanobodies for Diagnosis and Prognosis in Mouse Models of Liver Pathogenesis. Mol. Imaging Biol. 2017, 19, 49–58.
- Zheng, F.; Devoogdt, N.; Sparkes, A.; Morias, Y.; Abels, C.; Stijlemans, B.; Lahoutte, T.; Muyldermans, S.; De Baetselier, P.; Schoonooghe, S.; et al. Monitoring Liver Macrophages Using Nanobodies Targeting Vsig4: Concanavalin A Induced Acute Hepatitis as Paradigm. Immunobiology 2015, 220, 200–209.
