Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Virology
HIV-1 (human immunodeficiency virus type 1) infection begins with the attachment of the virion to a host cell by its envelope glycoprotein (Env), which subsequently induces fusion of viral and cell membranes to allow viral entry. Upon binding to primary receptor CD4 and coreceptor (e.g., chemokine receptor CCR5 or CXCR4), Env undergoes large conformational changes and unleashes its fusogenic potential to drive the membrane fusion.
- HIV
- envelope glycoprotein
- viral entry
- membrane fusion
1. HIV-1 Entry
The strategy that enveloped viruses, such as HIV-1 (human immunodeficiency virus type 1), use to gain entry into their host cells is membrane fusion, which is an energetically favorable process but with high kinetic barriers [1,2]. Virus-encoded fusion proteins are catalysts and undergo structural rearrangements from a high-energy, metastable prefusion conformation to a low-energy, stable postfusion conformation, providing free energy for overcoming these kinetic barriers [3,4,5]. In the case of HIV-1, its envelope glycoprotein (Env) functions as the fusion protein. The Env protein is synthesized as a precursor, gp160 (for glycoprotein with an apparent molecular weight of 160 kDa; Figure 1A), which forms a trimer (gp160)3 and is then cleaved by a cellular furin-like protease into two noncovalently associated subunits: the receptor-binding subunit gp120 and the fusion subunit gp41 [6]. Three copies of each subunit constitute the mature envelope spike (gp120/gp41)3. It is generally believed that sequential binding of gp120 to primary receptor CD4 and coreceptor (e.g., chemokine receptor CCR5 or CXCR4) initiates a cascade of refolding events in gp41 that drive the membrane fusion process [7,8]. The mature Env spikes are also the sole antigens on the surface of virion and induce strong immune responses in infected individuals [9,10]. Not surprisingly, HIV-1 Env is a critical target for the development of both vaccines and therapeutics against the virus. Recent advances in the structural biology of HIV-1 Env and its complexes with the host receptors, as well as in the design of novel fusion inhibitors, have provided new insights into HIV-1 entry and its inhibition. In this review, we summarize our latest understanding of the membrane fusion catalyzed by HIV-1 Env and discuss related therapeutic strategies to block viral entry.
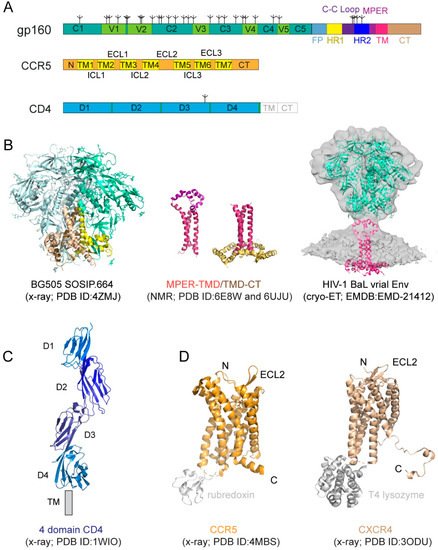
Figure 1. HIV-1 (human immunodeficiency virus type 1) envelope glycoprotein and its receptors. (A) The full-length HIV-1 Env, gp160. Segments of gp120 and gp41 include: C1–C5, conserved regions 1–5; V1–V5, variable regions 1–5; F, fusion peptide; HR1, heptad repeat 1; C-C loop, the immunodominant loop with a conserved disulfide; HR2, heptad repeat 2; MPER, membrane proximal external region; TM, transmembrane anchor; CT, cytoplasmic tail; tree-like symbols, glycans. Those for CCR5 include: N, N-terminus; TM1-7, transmembrane helices 1–7; ECL1-3, extracellular loop 1–3; ICL3, intracellular loop 1–3; and CT, cytoplasmic tail. For CD4, they are: D1–4, immunoglobulin (Ig) domain 1–4; TM and CT. (B) Structures of HIV-1 Env. The crystal structure of the unliganded HIV-1 BG505 SOSIP.664 Env trimer (pdb ID: 4ZMJ; [11]) that lacks the MPER, TMD, and CT is shown in the ribbon diagram with gp120 in cyan and gp41 in yellow. Structures of the MPER-TMD and TMD-CT reconstituted in bicelles that mimic lipid bilayer determined by NMR (pdb ID: 6E8W; [12]; pdb ID: 6UJU; [13]). The MPER is in magenta, the TMD in dark red, and the CT in gold. The EM density in gray is 3D reconstruction of the unliganded HIV-1 BaL Env spike on the surface of virion by cryo-electron tomography (EMDB ID: EMD-21412). (C) Crystal structure of soluble four domain CD4 (pdb ID: 1WIO; [14]). D1-D4 and the location of the transmembrane segment (TM) are indicated. (D) Crystal structure of a modified CCR5 in complex with Maraviroc (pdb ID: 4MBS; [15]). CCR5 is shown in the ribbon diagram in brown, the internally fused rubredoxin and the ligand in gray. N-terminus (N), C-terminus (C), and the second extracellular loop (ECL2) are indicated. Crystal structure of an engineered CXCR4 in complex with a viral chemokine antagonist IT1t (pdb ID: 3ODU; [16]). CXCR4 is shown in brown, the fused T4 lysozyme and the ligand in gray.
2. Structures of HIV-1 Env and Cellular Receptors
2.1. HIV-1 Env
The HIV-1 Env is a heavily glycosylated type I membrane protein with five conserved regions 1–5 (C1-C5) and five variable regions 1–5 (V1-V5) in gp120; and various segments in gp41 including a hydrophobic fusion peptide (FP), two heptad-repeat regions (HR1 and HR2), a disulfide-bridged loop (C-C loop), a tryptophan-rich membrane proximal external region (MPER), a transmembrane segment (TM) and a cytoplasmic tail (CT) (Figure 1A). The protein has been historically a very challenging target for structural analysis due to technical difficulties associated with large membrane-bound glycoproteins. Nevertheless, a truncated version of gp120, named ‘gp120 core’, with V1-V3 and terminal segments deleted, was crystallized in two forms: a deglycosylated one in complex with CD4 and a CD4-induced antibody for HIV-1 [17] and an unliganded and fully glycosylated one for closely related simian immunodeficiency virus (SIV) [18], producing structures that gave us the first glimpse of gp120 folding and its interaction with CD4. Likewise, the structure of a gp41 fragment of HR1 and HR2 has been solved by X-ray crystallography and nuclear magnetic resonance (NMR) [7,8,19,20,21], revealing the postfusion conformation of gp41 as a six-helix bundle, in which the HR1 and HR2 helices are arranged into a trimer of hairpins.
The first breakthrough on high-resolution structures of the Env trimer only came more than a decade later from a designed soluble construct, termed ‘SOSIP’, with stabilizing modifications (i.e., a disulfide bond between gp120 and gp41, an I559P substitution in gp41, and a truncation at residue 664 deleting the MPER; [22]) by both cryogenic electron microscopy (cryo-EM) and X-ray crystallography [23,24,25] (Figure 1B). Subsequently, the structure of a detergent-solubilized Env trimer without the CT and SOSIP modifications was determined in complex with neutralizing antibodies by cryo-EM [26]. More recently, the cryo-EM structures of two full-length HIV-1 Env constructs purified in detergent have also been reported [27,28]. These trimer structures have shown that the prefusion gp41 adopts a drastically different conformation from the postfusion six-helix bundle structure and provided much-needed insights on Env structure and its conformational changes. The MPER, TMD, and CT are all disordered in these structures, however, highlighting the important role of the lipid bilayer in stabilizing the structure of these regions. An attempt to determine the structure of the missing regions using a full-length Env reconstituted in lipid nanodiscs did not yield much additional high-resolution information [29]. In addition, cryo-electron tomography (cryo-ET) has been used to study the structures of Env trimer on the surface of both HIV and SIV chemically inactivated virions, leading to reconstructions at a low resolution (~20 Å) during early days [30,31,32,33], and a more recent one at ~10Å resolution (Figure 1B; [34]), but the regions near the membrane remain difficult to resolve.
Recent data indicate that the membrane-related components of HIV-1 Env, including the MPER, TM domain (TMD), and CT, influence the stability and antigenicity of the Env ectodomain, as well as cell–cell fusion and viral infection [12,35,36,37,38,39], in agreement with their conserved features. For example, the MPER has been studied extensively because it contains epitopes recognized by a group of broadly neutralizing antibodies [40,41,42,43,44]. The TMD has a ‘GXXXG’ motif and a highly conserved positively charged residue (Lys or Arg). The CT includes the Kennedy sequence, three conserved amphipathic α-helices segments referred to as a lentiviral lytic peptide (LLPs: LLP1, LLP2, and LLP3) [45,46,47]. Truncation of the CT of the full-length HIV-1 Envs has minimal impact on their fusogenic activity, but it has an unexpectedly large impact on the antigenic structure of the ectodomain [35]. Some other studies showed that the CT modifications had little effect on the Env antigenicity for certain HIV-1 isolates [48,49,50]. Nevertheless, structural studies in the context of a lipid bilayer appear to support crosstalk between the CT and the ectodomain.
NMR spectroscopy and isotopic labeling techniques enable the structural determination of peptides and small proteins at high resolutions, particularly for those with some flexibility. 2D Transverse relaxation-optimized spectroscopy (2D-TROSY) combined with 3D spectroscopies and selective deuterium-labeling methods allows structural characterization of oligomeric membrane proteins in micelles or bicelles. Moreover, functional dynamics and protein–protein/ligand interactions can be quantitatively measured by NMR techniques at the residue level. Thus, NMR spectroscopy is a powerful approach suitable for structural analysis of the MPER, TMD, and CT regions of gp41 [51]. For instance, the structure of the TMD reconstituted in bicelles mimicking a lipid bilayer was first determined by NMR [36] (Figure 1B). The TMD forms a well-ordered trimer, and that mutational changes disrupting the TMD trimer alter antibody sensitivity of the ectodomain, suggesting that the TMD contributes to Env stability and antigenicity. Moreover, although previous studies reported that the MPER might be buried in the viral membrane [52,53,54], the NMR structure that contains both the MPER and TMD in the bicelle system has shown that the MPER forms a well-ordered trimeric assembly, not buried in the membrane [12] (Figure 1B). The MPER mutations can destabilize the Env ectodomain and shift it towards an open conformation, suggesting that the MPER is a control relay that modulates open and closed states of the Env trimer. Furthermore, a third NMR structure containing the TMD and part of CT in bicelles has been reported recently [13] (Figure 1B), revealing that the CT folds into amphipathic helices, attached to the bilayer surface, wrapping around the C-terminal end of the TMD and thereby forming a support baseplate for the entire Env. Mutational data have also confirmed that altering the CT-TMD packing interface affects the antigenicity of the full-length Env trimer. These results support the model of a dynamic coupling across the TMD between the CT and ectodomain. Nevertheless, a high-resolution structure of the native, full-length HIV-1 Env in the membrane is still required for a full understanding of its structure and function.
2.2. Primary Receptor CD4
CD4 (cluster of differentiation 4) is a membrane-bound glycoprotein found on the surface of immune cells and normally functions as a coreceptor for enhancing T-cell receptor-mediated signaling. CD4 contains four immunoglobulin-like extracellular domains, D1–D4 (Figure 1A), exposed on the cell surface. It was shown to be the primary HIV-1 receptor shortly after the discovery of the HIV virus [55,56,57]. The structure of CD4 alone or in complex with gp120 core has been determined by X-ray crystallography [14,17,58,59,60] (Figure 1C).
2.3. Coreceptor
CD4 alone was not sufficient to support HIV-1 infection, leading to intensive search and subsequent identification of CXCR4 and CCR5, the seven-transmembrane (7TM) chemokine receptors, as the coreceptor for the virus [61,62,63,64,65,66]. Coreceptor usage is the primary determinant for viral tropism [67], as those that use CCR5 (R5 viruses) are the dominant form during viral transmission, and others using CXCR4 (X4 viruses) or both (dual-tropic; R5/X4 viruses) emerge mainly during disease progression [68,69,70,71]. Both CCR5 and CXCR4 have a core structure formed by 7TM helices, decorated by an N-terminal segment and three extracellular loops (ECL) exposed outside of the cell, as well as three intracellular loops (ICL), and a cytoplasmic C-terminal tail on the opposite side of the membrane (Figure 1A). A C-terminally truncated CXCR4 construct with stabilizing mutations, and a T4 lysozyme fusion in complex with different ligands, and a similarly modified CCR5 construct containing a rubredoxin fusion in complex with the anti-HIV drug, Maraviroc, have been crystallized, yielding high-resolution structures [15,16,72] (Figure 1D), with a typical 7TM helical bundle topology typically seen for other GPCRs (Figure 1C; [73]). A two-site model has been proposed for their ligand interactions [74], as the N-terminal segment of CXCR4 or CCR5 forms the chemokine recognition site 1 (CRS1) to bind the globular core domain of chemokine, while their TM helices make up the chemokine recognition site 2 (CRS2) to interact with the N-terminus of the chemokine. These structures have revealed the general architecture of these chemokine receptors and their interactions with the ligands [15,16,72,75], but they did not provide many of the molecular details of how they function as HIV-1 coreceptors.
3. Molecular Mechanism of HIV-1 Membrane Fusion
3.1. Interactions between HIV-1 Env and Cellular Receptors
3.1.1. Interactions between Env and CD4
The binding affinity is in the low nM range for soluble CD4 and monomeric gp120 [76,77,78], but it can be ~20 nM for various trimeric forms of soluble Env trimers [78,79]. The binding interface between CD4 and gp120 was first defined in the structure of the gp120 core-CD4 complex [17]. Gp120 core has two separate domains—inner domain and outer domain, and there is also a four-strand β-sheet, named bridging sheet, between the two domains. CD4 interacts with gp120 mainly at the interface between the inner domain and outer domain, inducing the formation of the bridging sheet. The structures of CD4 in complex with Env trimer have been determined using the SOSIP trimer as well. The CD4 bound trimer adopts a more open conformation compared with the unliganded Env SOSIP trimer (Figure 2A). The Env conformational changes include V1–V2 flip, V3 exposure, the bridging sheet formation, and repositioning of the fusion peptide in gp41 [80,81]. Another further constrained trimer, named DS-SOSIP.664, was created by introducing a disulfide bond (201C–433C) into the SOSIP [11]. This trimer binds sCD4 with an asymmetric 1:1 (CD4:trimer) stoichiometry. It appears that a single CD4 molecule is embraced by a quaternary HIV-1 Env surface with the previous defined CD4-binding region in the outer domain of one gp120 protomer and with the second CD4-binding site (CD4-BS2) in the inner domain of a neighboring gp120 protomer (Figure 2A), suggesting that the complex may represent the initial contact of the HIV-1 Env with the CD4 receptor [82].
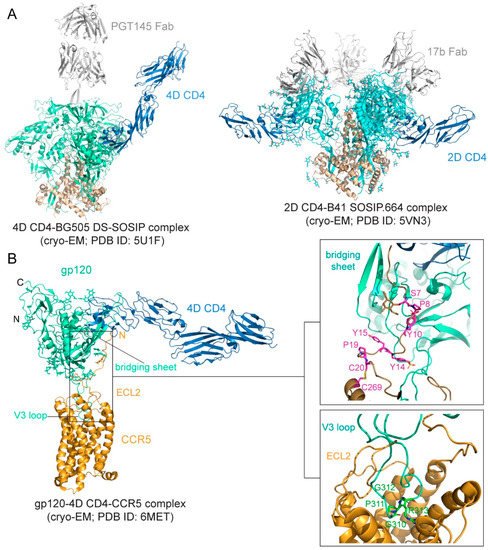
Figure 2. Env-CD4 interaction. (A) Left, the cryo-EM (cryogenic electron microscopy) structure BG505 DS-SOSIP.664 Env trimer in complex with 4D CD4 and PGT145 Fab (pdb ID: 5U1F; [82]) is shown with gp120 in cyan, gp41 in brown, CD4 in blue and PGT145 Fab in gray. Right, the cryo-EM structure of B41 SOSIP.664 Env trimer in complex with 2D CD4 and 17b (pdb ID: 5VN3; [80]) is shown with gp120 in cyan, gp41 in brown, CD4 in blue, and 17b Fab in gray. (B) Env-CCR5 interaction. Left, overall structure of the 4D CD4-gp120-CCR5 complex (pdb ID: 6MET; [83]) shown in ribbon diagram. N, N-terminus; C, C-terminus; ECL2, extracellular loop 2. V3 loop and the bridging sheet of gp120 are also indicated. Right, close-up views of the interfaces between gp120 and CCR5. The N-terminus of CCR5 is attaching to the surface of the four-stranded bridging β sheet formed by the V1V2 stem and β21–β22 of gp120. Residues Ser7, Pro 8, sulfated Tyr 10, sulfated Tyr14, Tyr15, and Pro19, as well as the disulfide between Cys20 and Cys269 of CCR5 are highlighted in the stick model. The O-linked glycan at Ser7 is also shown. V3 is inserting into the CRS2 of CCR5. The conserved GPGR motif of V3 is highlighted in the stick model, and ECL2 of CCR5 is indicated.
3.1.2. Interactions between Env and Coreceptor
Preparing stable and homogenous samples of purified CCR5 or CXCR4 has been technically challenging, and various assays have, therefore, been employed to measure the binding affinity for the Env-coreceptor interactions in the presence of soluble CD4 (<10 nM for CCR5; 200–500 nM for CXCR4; [84,85,86,87]). The first structure of a full-length monomeric gp120 in complex with a soluble 4D-CD4 and an unmodified human CCR5 was determined by cryo-EM [83] (Figure 2B), revealing details of the interactions between gp120 and CCR5, largely consistent with the predictions based on previous mutational data [88,89]. The crown of the V3 loop insets into a deep pocket formed by the 7-TM helices of CCR5. The ECL2 forms a semicircular grip and wraps around the V3 loop, making contact with residues from both the V3 stem and crown. The N terminus of CCR5 and the bridging sheet of gp120 make up the second interface between them, in which the N terminal segment of the coreceptor adopts an extended conformation with several sharp turns and latches onto the surface of the bridging sheet (Figure 2B).
3.2. Membrane Fusion
Previous studies suggested a working model for HIV-1 Env-mediated membrane fusion [7], depicted in Figure 3. Gp120 binding to the receptors induces large structural rearrangements in gp41, which adopts a prefusion conformation within the precursor gp160. The cleavage between gp120 and gp41 primes the protein and makes it metastable with respect to the postfusion conformation. Once triggered, the FP of gp41 translocates and inserts into the target cell membrane. Further refolding of gp41 into a hairpin conformation creates a six-helix bundle structure, thereby placing the FP and TM segments at the same end of the molecule and effectively bringing the two membranes together. Formation of hemifusion stalk and fusion pore ensues membrane fusion and entry of the viral capsid into the target cell. How many receptor molecules are necessary for activating one Env trimer and how many Env trimers are required to induce productive viral entry remain controversial [90]. Early data demonstrated that a fully functional Env trimer could be assembled even with one or more gp120 protomer(s) defective in CD4 binding or coreceptor binding [91], suggesting that a single CD4 or coreceptor is sufficient to trigger the Env trimer. Similarly mixed trimer assays have been used to estimate the number of Env trimers needed for membrane fusion by mathematical modeling, giving a range between 1 and 8 (reviewed in [90]). More recently, an intriguing stoichiometry among Env trimer, CD4, and coreceptor CCR5 or CXCR4 during membrane fusion has been estimated by advanced microscopy and spectroscopy imaging techniques and involves oligomerization of the receptors [92]. smFRET (single molecule fluorescence resonance energy transfer) data also suggest that one CD4 could induce large structural changes within the Env trimer [93]. In another study, fusion inhibitors have been used to trap the pre-hairpin intermediate state of gp41 in an extended conformation at the viral attachment sites, followed by visualization of electron tomography, showing only 2–3 gp41 molecules per attached virion [94]. Since there are only ~14 Env spikes per virion [32], the probability for multiple Env trimers to engage multiple CD4 and coreceptor molecules simultaneously in a synchronized manner would low. Nevertheless, recent progress in the structural biology of HIV-1 Env and its complexes with the cellular receptors has allowed us to fill in additional molecular details of this working model.
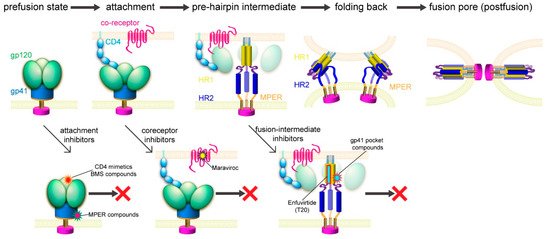
Figure 3. HIV-1 membrane fusion and its inhibition. Top, membrane fusion likely proceeds stepwise as follows. (1) Binding of gp120 to CD4 and a coreceptor allows viral attachment and triggers structural changes in Env. (2) Dissociation of gp120 and insertion of the fusion peptide of gp41 into the target cell membrane leads to the prehairpin intermediate [95]. (3) HR2 folds back onto the inner core of HR1 and brings the two membranes together. (4) A hemifusion stalk forms and resolves into a fusion pore [96]. Bottom, opportunities for fusion inhibitors, including attachment inhibitors targeting the CD4 binding site and the MPER; coreceptor inhibitors; and fusion-intermediate inhibitors.
The binding of gp120 to CD4 attaches the virus to the surface of the target cell. The SOSIP trimer in a complex of one CD4 molecule shows a possible orientation for CD4 to make the first contact with the Env (Figure 2A). It has been long speculated that the coreceptor binding induces additional structural changes in gp120 that lead to the irreversible refolding of gp41. Unexpectedly, the CD4-gp120-CCR5 structure revealed no obvious allosteric changes in gp120 that could propagate from the CCR5 binding site to gp41, as a comparison of the CD4- and CCR5-bound gp120 and the CD4-bound gp120 showed no major differences in the gp120 core region ([83]; Figure 2B). The only obvious structural changes were the reconfiguration of the V3 loop and flipping back of the N- and C-termini of gp120 near its interface with gp41. In the prefusion structure of the Env trimer [23,24,25,26,80], gp41 folds into a so-called “4-helix collar” with its four helices [25], wrapping around the N- and C-termini of gp120. If gp120 departures, gp41 would be destabilized and likely enter an irreversible refolding process. Partial or complete gp120 dissociation may, therefore, be the crucial “trigger” that prompts a series of refolding events in gp41 and the membrane fusion process. Indeed, CD4 binding causes a large shift of the C-terminal helix away from the gp120 termini, creating a pocket filled by the fusion peptide [80], which normally packs against the gp120 N-terminus. When the fusion peptide flips away from the pocket because of the intrinsic conformational dynamics, it opens up one side of the gp41 grip on the gp120 termini, and the N-terminal segment of gp120 can then bend back to adopt the conformation seen in the CCR5-bound structure. The replacement of the gp120 termini can prevent the fusion peptide from reoccupying the pocket and effectively weaken the gp120-gp41 association. Spontaneous or CD4-induced gp120 dissociation from the Env trimers has been well documented for many HIV-1 isolates [97,98], indicating that gp120 has the tendency to dissociate from gp41 even without a coreceptor. We note that this model is very similar to that proposed for membrane fusion catalyzed by coronavirus spike proteins, in which dissociation of the receptor-binding subunit initiates the irreversible refolding of the metastable fusion subunit, allowing the fusogenic transition to a stable postfusion structure [99,100].
If CCR5 does not induce further structural rearrangements in gp120 to activate gp41, what is a coreceptor needed for then? First, it would be non-productive if gp120 dissociates prematurely in the absence of a coreceptor, because when a virion attaches to the target cell surfaces with the Env trimer forming a complex only with CD4, the distance between the gp41 fusion peptide and the cell surface is not close enough for it to reach the target membrane. Engaging a coreceptor, which is largely embedded in the membrane, will bring the fusion peptide substantially closer [25]. Second, single-molecule force spectroscopy data using infectious virions and live host cells indicated that the Env-CD4 association is unstable and rapidly reversible unless CCR5 binding immediately follows [101,102]. CCR5 is needed merely to stabilize the CD4-induced conformational changes in Env, which are already sufficient to drive membrane fusion. Third, fusion pore formation probably requires 2–3 Env trimers clustered together [26,90,94], also demonstrated for other viral fusion proteins [103]. A stable Env-receptor complex would help synchronize these trimers to undergo the same conformational changes. Thus, a coreceptor probably functions by stabilizing and anchoring the CD4-induced conformation of the Env trimer near the cell membrane to facilitate productive membrane fusion.
This entry is adapted from the peer-reviewed paper 10.3390/v13050735
This entry is offline, you can click here to edit this entry!
