Adenosine diphosphate (ADP)-ribosylation is a nicotinamide adenine dinucleotide (NAD+)-dependent post-translational modification that is found on proteins as well as on nucleic acids. Promising technological advances have enabled the development of innovative tools to detect NAD+ and NAD+/NADH (H for hydrogen) ratios as well as ADP-ribosylation. These tools have significantly enhanced our current understanding of how intracellular NAD dynamics contribute to the regulation of ADP-ribosylation as well as to how mono-ADP-ribosylation integrates into various cellular processes.
- NAD
- NADH
- ADP-ribose
- ADP-ribosylation
- MARylation
- PARylation
- ART
- PARP
- competition
- cancer
- immunofluorescence
- macrodomain
- Af1521
1. ADP-ribosylation (Introduction)
ADP-ribosylation (ADPR) is a covalent chemical modification conserved throughout all domains of life except budding yeast [1]. The modification was initially identified as a post-translational protein modification, though it has recently been found on nucleic acids as well [2,3,4,5,6]. ADP-ribosylation is catalyzed by ADP-ribosyltransferases (ARTs) and consists of the transfer of ADP-ribose (ADPr) from nicotinamide adenine dinucleotide (NAD+) onto the substrate with subsequent release of nicotinamide (NAM) [7,8,9]. NAD+ and its reduced form NADH are both important redox equivalents and key cofactors for the electron transport chain, thereby fueling oxidative phosphorylation (OXPHOS) [10,11]. Therefore, the dependence of ADP-ribosylation on NAD+ directly links the modification to cell metabolism. ADP-ribosylation comes in two different flavors: while the attachment of a single ADP-ribose molecule is referred to as mono-ADP-ribosylation (MARylation), the gradual attachments of multiple ADP-ribose moieties onto one another results in oligo or poly-ADP-ribosylation (PARylation) [8,12].
Mammalian ARTs (i.e., writers) have classically been divided into three groups: (i) clostridium toxin-like ARTs (ARTCs) are mainly described to catalyze extracellular ADP-ribosylation, while (ii) diphtheria toxin-like ARTs (ARTDs or poly-ADP-ribose polymerases (PARPs)) and (iii) sirtuins Sirt 4, 6 and 7 catalyze ADP-ribosylation in different intracellular compartments [12,13,14]. Among those enzymes, only four proteins (ARTD1/PARP1, ARTD2/PARP2 and ARTD5 and 6/tankyrase 1 and 2) have been reported to possess PARylation activity [12]. With the exception of ARTD13/PARP13, whose activity could not yet be detected, all remaining intra- and extracellular ARTs and sirtuins catalyze MARylation [15] (Figure 1).
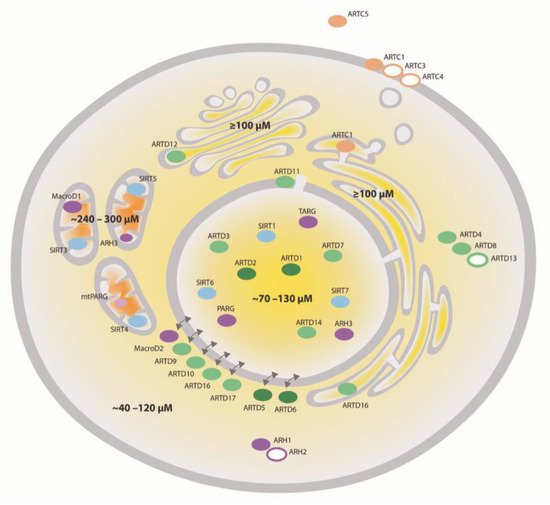
Figure 1. Compartmentalization of NAD+-converting enzymes and NAD+. NAD+-converting enzymes have been identified in different cellular compartments. ARTDs are colored in green, ARTCs—in beige, sirtuins—in blue and ADP-ribosylhydrolases (ARHs)—in purple. Filled circles symbolize active enzymes while open circles indicate enzymes whose activity has not been confirmed so far. The intensity of the color shows the expected intensity of the enzymatic activity. The concentration of NAD+ (depicted in different yellow to orange shades) is high in mitochondria (∼300 μM), intermediate in the nucleus and cytosol (∼100 μM) and low (<1 μM) in the extracellular space and can vary considerably depending on the cell type, metabolic condition, stress and redox status. Poly-ADP-ribose glycohydrolase (PARG); Terminal ADP-ribose protein glycohydrolase (TARG).
In addition to these enzymes, recent studies have proposed that other protein families, including NEURL4-like enzymes and certain leucine-rich repeat-containing enzymes, are able to catalyze ADP-ribosylation as well [16,17]. In recent years, protein ADP-ribosylation has emerged as a complex and dynamic post-translational modification that either directly affects the modified proteins or leads to modification-dependent protein complex formation thus serving as a signaling molecule [18]. ADP-ribosylation-dependent signaling often involves so-called readers―proteins that contain specific domains able to bind the mono-ADP-ribose (MAR) and/or the poly-ADP-ribose (PAR) [12,19]. Representative domains such as macrodomains, WWE domains or PAR-binding zinc fingers selectively recognize different forms of ADP-ribose such as MAR, PAR and oligo-ADP-ribosylation.
ADP-ribosylation is believed to be a reversible modification. The removal of ADP-ribosylation is mediated by ADP-ribosylhydrolases (ARHs, e.g., eraser) that either hydrolyze PAR or MAR or both [20,21,22,23,24]. MAR-specific ARHs seem to function in an ADPr amino acid acceptor site-specific manner [25]. The mammalian ARHs can be classified into ADP-ribosyl glycohydrolases and macrodomain-containing hydrolases, including their most prominent member, poly-ADP-ribose glycohydrolase (PARG), and its different isoforms [12]. As for the writers of ADP-ribosylation, the erasers also localize throughout the whole cell, such that together they cover every subcellular compartment (Figure 1). Amongst the ARHs, ARH1 and the inactive ARH2 localize to the cytoplasm, while ARH3 was shown to be nuclear and partially mitochondrial [26]. The macrodomain-containing ARH MacroD1 was shown to be mainly mitochondrial, while MacroD2 is cytoplasmic and TARG localizes to the nucleus [27,28,29]. Depending on the isoform, PARG, the major PAR degrading enzyme, either localizes to the nucleus, the cytoplasm or mitochondria [30]. Interestingly, MAR and PAR are also in vitro substrates for Nudix hydrolases and phosphodiesterases, although in this reaction, the modification is not completely removed, generating phosphoribosyl-modified proteins [31,32].
Although writers, readers and erasers of intracellular mono-ADP-ribosylation have been identified only recently, it is becoming more and more evident that this reversible post-translational modification is involved in a plethora of physiological and pathophysiological processes. More specifically, the modification plays vital roles in regulating cellular stress responses related to the quality control of DNA, RNA and proteins, such as the DNA damage response and the cytoplasmic stress response [1]. In addition, ADP-ribosylation is involved in host–pathogen interactions and several inflammatory signaling pathways [9,33]. In non-stress and non-self-defense pathways, ADP-ribosylation contributes to the regulation of cell cycle progression, gene expression, telomere length and protein stability [34]. Finally, ADP-ribosylation has also been shown to favor pathophysiological conditions such as tumorigenesis and tumor progression, in which it has been described to regulate the unfolded protein response (UPR) [35], the cytoplasmic stress response, miRNA-mediated post-transcriptional gene regulation [36], cancer-related signal transduction pathways or cell migration [37].
2. NAD+ Synthesis
Since NAD+ is the only known ADP-ribose donor, ADP-ribosylation is tightly linked to the availability and subcellular distribution of NAD+ pools. In fact, in cell culture, the turnover of ADP-ribosylation was found to directly correlate with overall NAD+ levels and NAD+ synthesis [38]. NAD+ availability depends on two factors: synthesis capacity and the cellular redox state. In cells, NAD+ can be synthesized de novo from tryptophan, via the Preiss–Handler pathway from nicotinic acid or from its breakdown products nicotinamide or nicotinamide riboside (NR) via the so called salvage pathway [39,40,41]. The salvage pathway is especially important for the restoration of intracellular NAD+ pools following extensive enzymatic consumption, e.g., upon hyperactivation of ARTD1 and subsequent hyper-consumption of NAD+. The choice of NAD+ synthesis pathways depends on the expression pattern of the respective enzymes involved in either pathway and was shown to be highly cell type- and organ-specific. With the exception of the liver where NAD+ levels were shown to predominantly depend on tryptophan, many tissues and most transformed cell culture cell lines synthesize NAD+ from NAM and thus rely mostly on the salvage pathway [38]. The salvage pathway depends on the expression level of nicotinamide phosphoribosyltransferase (NAMPT) and the nicotinamide mononucleotide adenylyl transferases (NMNATs), NMNAT1, 2 and 3 [42,43]. The localization of the three NMNATs is described to be enzyme-specific, with NNMAT1 localizing mainly to the nucleus, NMNAT2—to the cytoplasm as well as to the Golgi apparatus and NMNAT3—to the mitochondria [44,45,46]. While NAMPT is mostly found in the nucleus and the cytoplasm, a small fraction is believed to be mitochondrial, thus enabling all three compartments to fully resynthesize NAD+ from NAM. Given that many small metabolites are believed to freely diffuse between the nucleus and the cytoplasm, nuclear and cytoplasmic NAD+ pools are thought to be interconnected. In line with that, in certain cell culture settings, NMNAT1 can compensate for the lack of NMNAT2 and vice versa. In strong contrast to those in vitro experiments, knockout of either NMNAT1 or NMNAT2 in mice is lethal [47,48]. Hence, although NAD+ is thought to freely shuttle between the nucleus and the cytoplasm in vivo, neither enzyme seems to be able to functionally compensate for the loss of the respective other one. Comparably, mutations in either the NMNAT1- or the NMNAT2-encoding gene in humans were shown to result in the early development of severe disorders [49,50], again suggesting that the regulation of these proteins is compartmentalized and non-redundant in vivo. A comprehensive overview about recent advances in understanding NAD+ metabolism and it impact on health and disease is reviewed elsewhere [1].
3. Regulation and function of Cellular Mono-ADP-ribosylation
The establishment of protein MARylation can be regulated on various levels. First, expression and cellular distribution of enzymes largely contribute to controlling the abundance of the enzymes, their localization and their target spectrum. Second, the ADP-ribosylation ability of every ART is naturally linked to the availability of NAD+ and its Km for NAD+ that varies from enzyme to enzyme and ranges from only few µM to several hundred µM [93,94,95,96]. Finally, comparable to other enzymes, mono-ARTs can be regulated by additional signaling events and binding to cofactors ([35,97,98] and reviewed in [99,100]).
4. Detection of Mono-ADP-Ribosylation
The two main challenges for the development of ADP-ribosylation detection tools can be attributed to the heterogenic nature of ADP-ribosylation (MARylation vs. PARylation) and similarity of this molecule to several other abundant cellular biological (macro-)molecules, including adenine nucleotides and nucleic acids [156,157]. Many antibodies against poly-ADP-ribose can thus recognize RNA and DNA and vice versa [158,159,160]. The heterogeneity can be attributed to the fact that ADP-ribosylation is catalyzed by at least 25 distinct enzymes with cell type-specific expression patterns and different cellular localizations and targets, as well as amino acid preferences. The currently available ADP-ribosylation tools can roughly be divided into three different categories, which are summarized in Table 2: (i) anti-ADP-ribose or PAR-binding antibodies, (ii) ADP-ribose-binding domains and (iii) chemical labeling of ADP-ribose. All three approaches and the most recent advances made in each category are discussed in the following sections.
Table 2. Overview of currently available ADP-ribosylation detection tools.
|
Antibodies |
|||
|
Type of ADPR |
Specificity |
Potential Application and Caveats |
Reference |
|
PAR |
PAR |
Immunfluorescence (IF), Western blotting (WB) for PAR Unable to detect oligomers and MAR |
[161] |
|
Pan-ADP-ribose |
MAR, PAR, amino acid-independent |
IF, WB for MAR and PAR Compatible with pulldowns and MS Cross-reactivity with nucleotide derivatives and ATP-derived PTMs possible Might have biases towards specific ADPr amino acid acceptor sides |
|
|
MAR |
MAR, amino acid-independent |
IF, WB for aa- and protein-specific MARylation Potentially compatible with pulldowns and MS |
[148] |
|
MAR attached to Ser or Thr |
IF, WB for aa- and protein-specific MARylation Potentially compatible with pulldowns and MS |
[148] |
|
|
ADP-Ribose-Binding Domains |
|||
|
Type |
Specificity |
Potential Application and Caveats |
Reference |
|
WWE |
PAR, oligo ADPR |
IF, WB, chromatin affinity precipitation (ChAP) for PAR Cross-reactivity with nucleic acids possible |
|
|
Macro(H2A1.1) |
PAR, MAR |
IF, WB for PAR and MAR Might have a bias towards specific ADPR types |
[162] |
|
Macro(Af1521) |
PAR, oligo, MAR |
IF, WB, ChAP for PAR Compatible with pulldown methods (Immunoprecipitation (IP), MS) Might have a bias towards specific ADPR types. |
|
|
Macro3x (ARTD8/PARP14) |
PAR (faint), MAR |
IF, WB for PAR Compatible with pulldown methods (IP). Fusion to green fluorescence protein (GFP) allows real-time analyses Might have a bias towards specific ADPR types |
|
|
Chemical Labeling |
|||
|
Type |
Specificity |
Potential Application and Caveats |
Reference |
|
Enzymatic labeling of terminal ADP-ribose (ELTA) |
PAR, oligo, MAR |
Compatible with pulldown methods (IP, MS) Can differentiate between different chain length |
[166] |
|
NAD labeling |
PAR, MAR |
Compatible with pulldown methods (IP, MS) |
|
This entry is adapted from the peer-reviewed paper 10.3390/cells10030680
References
- Claudia C.S. Chini; Julianna D. Zeidler; Sonu Kashyap; Gina Warner; Eduardo Nunes Chini; Evolving concepts in NAD+ metabolism. Cell Metabolism 2021, 33, 1076-1087, 10.1016/j.cmet.2021.04.003.
