Callose is a β-(1,3)-D-glucan polysaccharide with some β-1,6-branches that exists in all multicellular green algae and higher plants.
- callose
- papillae
- PMR4
- plant cell wall defense
1. Introduction
Callose is a β-(1,3)-D-glucan polysaccharide with some β-1,6-branches that exists in all multicellular green algae and higher plants [1]. In most plants, callose is synthesized by a family of callose synthases and plays important roles in several important biological processes in the plant. During cell division, callose is transiently deposited in the cell plate of the cells undergoing cytokinesis [2][3]. Callose is also a critical component of the transient cell wall surrounding pollen mother cells, the four microspores after meiosis and the pre-cell wall of the growing pollen tube tip [1][4]. Callose is also present in sieve plates, a basic component of the phloem, under normal growing and developmental conditions and can accumulate rapidly and plugs the sieve pores when subjected to stress [5][6]. Similar to this stress response, callose biosynthesis and degradation in the neck region of plasmodesmata help to regulate permeability during abiotic and biotic stresses [7]. In addition, callose is deposited between the plasma membrane and the pre-existing cell wall at sites of pathogen attack [8]. This pathogen-induced callose deposition functions as a chemical and physical defense mechanism for reinforcing plant cell wall and plays an essential role in the defense response to invading pathogens. Over the past 20 years or so, a large number of studies have been published on defense-related callose deposition that address the dynamic nature of pathogen-induced callose deposition, its important roles in plant defense responses, and the complex regulatory mechanisms of defense-related callose deposition.
2. Defense-Related Callose Deposition in Plants
Plants are constantly exposed to a large number of microbial pathogens and have evolved complex disease resistance mechanisms that protect plants from pathogens. The first common mechanism of plant disease resistance is through pre-formed structures and compounds such as plant cuticle surfaces, plant cell walls, antimicrobial chemicals and peptides, and other arsenals that inhibit or block pathogen-derived toxins, enzymes, or other activities [9]. The second common disease resistance mechanism is an infection-induced response of the interconnected two-tier immunity system [10]. The first layer is triggered upon recognition of pathogen-associated molecular patterns (PAMPs) by plant plasma membrane-localized pattern recognition receptors (PRRs). Well-characterized PRRs include Arabidopsis FLS2 (Flagellin-sensing 2) and EFR (the EF-Tu receptor), which recognize bacterial flagellin and the elongation factor-Tu (EF-Tu), respectively [11]. Adapted pathogens deliver effector proteins to plant cells to suppress PAMP-triggered immunity (PTI) [12]. To counter this, plants have evolved the second layer of the immunity system through recognition of pathogen effectors to activate effector-triggered immunity (ETI). Receptors for ETI are often plant resistance (R) proteins containing nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains with a Toll-interleukin receptor (TIR) or coiled-coil (CC) N-terminal domain [13]. Both PTI and ETI are associated with diverse signaling processes including dynamic protein interactions and phosphorylation, generation of reactive oxygen species (ROS), Ca++ signal spike, and mitogen-activated protein kinase (MAPK) activation [14]. These signaling processes then converge in the nucleus to elicit host transcriptional reprogramming not only for defense but also for balancing defense with growth [15].
In Arabidopsis, there are 12 genes encoding GLUCAN SYNTHASE-LIKE (GSL) callose synthases, and pathogen- and PAMP-induced callose deposition is dependent on the GSL5/(POWDERY MILDEW RESISTANT 4 (PMR4) callose synthase [16]. Callose-induced PAMPs include the 22–amino acid sequence of the conserved N-terminal part of flagellin (Flg22) and the 18 amino acid peptide of the bacterial elongation factor EF-Tu (Elf18) [17]. Fungal cell elicitors including chitin, a β-(1,4)-linked polymer of N-acetylglucosamine, and chitosan, a randomly distributed β-(1,4)-linked polymer of D-glucosamide and acetylglucosamine, are also potent PAMPs for inducing callose deposition [18]. Besides PAMPs, endogenous damage-associated patterns (DAMP) such as oligogalacturonides from pathogen- or herbivore-damaged plant tissues can function as elicitors to activate callose depositions as well [19]. Pathogen-induced callose can also be primed or enhanced by prior pathogen infection or defense-inducing compounds. For example, induction of systemic acquired resistance (SAR) by local pathogen infection is associated with augmented levels of callose upon secondary pathogen inoculation [20]. Furthermore, resistance-inducing chemicals including salicylic acid (SA), SA analog benzo(1,2,3) thiadiazole-7-carbothioic acid S-methyl ester (BTH), and the nonprotein amino acid BABA can augment depositions of pathogen-inducible callose [21][22]. It has also been recently reported that in tomato and wheat callose priming can be induced by Rhizophaus irregularis, a mycorrhizal fungus that can enhance resistance to several pathogens such as the necrotrophic fungal pathogen Botrytis cinerea in tomato [23][24].
One of the earliest plant defense responses against haustorium-forming pathogens is the deposition of a cell wall- associated apposition called papillae for halting the fungal penetration attempt [8]. If the fungal pathogen is successful in penetration and forms a haustorial feeding structure, the cell wall apposition materials can form collars or neck bands, including partial or even full encasements around the haustorium [25]. Common biochemical constituents of papillae include callose, phenolic, and phenolic conjugates such as phenlic-polyamines, ROS, peroxidases, cell wall structural proteins such as arabinogalactan proteins and hydroxyproline-rich glycoproteins, and cell wall polymers including pectin and xyloglucans [25]. Immuno-fluorescence and immune-gold labeling also found that Arabidopsis encasements surrounding the haustoria of the powdery mildew fungal pathogen Golovinomyces orontii contain callose, arabinogalactan proteins, rhamnogalacturonanI, a β-linked galactose-containing protein, and xyloglucanin [26], supporting the idea that the papillae and encasements are related and their formation is associated with increased callose deposition.
Callose deposition is also induced as a defense mechanism in cell wall near the neck zone of plasmodesmata to control their permeability [27]. Plasmodesmata are symplastic junctions between cells that function as important pathways for intercellular communication and molecular exchange, including cell-to-cell spread of viruses [28][29]. The level of callose in the plasmodesmatal neck zone is important for the cell-to-cell movement of cellular molecules and viruses; high levels of callose reduce or even close plasmodesmatal channels, while low levels of callose open them [28][29] (Figure 1). The level of callose in the plasmodesmata is regulated by two groups of enzymes; callose synthases synthesize callose while β-1,3-glucanases degrade it [28][29]. Callose accumulation in the plasmodesmata is induced by many viruses including Tobacco mosaic virus (TMV), Maize dwarf mosaic virus (MDMV), Potato virus X (PVX), and Tomato bushy stunt virus (TBSV) [7][28][29][30].
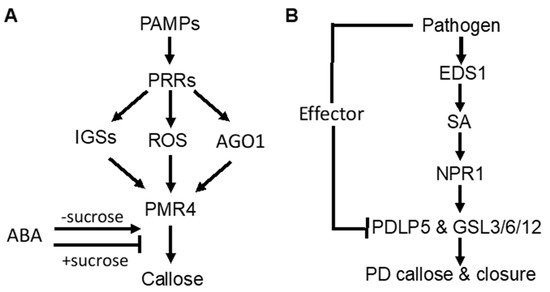
Figure 1. Signaling of pathogen-induced callose deposition in plants. (A) pathogen-associated molecular pattern (PAMP)-induced callose deposition is mediated by pattern-recognition receptors (PRRs) and promoted by indole glucosinolates (IGSs), reactive oxygen species (ROS), and RNAi regulatory protein Argonaute1 (AGO1). ABA stimulates but represses PAMP-induced callose deposition in the presence of high sucrose concentrations. (B) pathogeninduced callose deposition in plasmodesmata (PD) through the SA-mediated signaling pathway.
3. Regulation of Defense-Related Callose Deposition
3.1. Signaling Pathways Controlling Pathogen-Induced Callose Deposition
As part of PTI, PAMP-induced callose deposition is under the control of plant PRRs. Activity of the downstream pathways is marked by common signaling events, such as anion fluxes, protein phosphorylation cascades, accumulation of ROS, and defense gene induction. In Arabidopsis, ROS act as positive signals in Flg22-induced callose [31][32] (Figure 1A). The RNAi regulatory protein Argonaute1 also generates various miRNA signals that stimulate or repress Flg22-induced callose [33] (Figure 1A). Flg22-induced callose in Arabidopsis also requires intact biosynthesis of 4-methoxylated indole glucosinolates [34] (Figure 1A), suggesting that these secondary metabolites or break-down products play a crucial role in the regulation of callose. Interestingly, Flg22- and chitosan-induced callose differ in the requirement for the NADPH oxidase RBOHD, the glucosinolate regulatory enzymes PEN2, and the callose synthase PMR4 [35]. The rbohD mutant accumulated reduced levels of Flg22-induced H2O2 and failed to deposit enhanced levels of callose upon treatment with Flg22. However, the rbohD mutant deposited normal levels of callose in response to chitosan, even though chitosan-induced H2O2 was reduced. Likewise, the pen2-2 mutant failed to deposit enhanced callose in response to Flg22, whereas chitosan induced statistically significant enhancements in callose deposition. The pmr4-1 mutant deposited dramatically reduced levels of basal callose and failed to respond to Flg22. However, chitosan still elicited a residual callose response in pmr4-1 plants, indicating that one or more other callose synthases than PMR4 are responsible for a portion of chitosan-induced callose. These results demonstrated that PAMP-induced callose is controlled by distinct signaling pathways.
SA plays an important role in pathogen-induced plasmodesmata closure and associated callose deposition in Arabidopsis [36]. Direct exogenous application of SA or bacterial infection induce plasmodesmata closure and callose deposition. SA pathway mutants are impaired in this response. The SA- or pathogen-induced plasmodesmata closure and associated callose deposition requires an ENHANCED DESEASE RESISTANCE1(EDS1)– and NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1)–dependent SA pathway and is dependent on the regulator of plasmodesmal gating PLASMODESMATA-LOCATED PROTEIN5(PDL5) [36](Figure 1B). These results indicate that SA is a crucial signal in the regulation of cell-to-cell communication and trafficking via plasmodesmata in plant cells during innate immune responses.
Abscisic acid (ABA) signaling has an important effect on induced or primed callose deposition. Callose priming induced by the nonprotein amino acid β-amino butyric acid (BABA) requires an intact ABA-dependent pathway in Arabidopsis. BABA-induced resistance in Arabidopsis against Plectosphaerella cucumerina is known to be mediated by callose priming. Indole-3-carboxylic acid (ICOOH, also known as I3CA) mediates BABA-induced resistance in Arabidopsis against P. cucumerina. I3CA treatment increased ABA levels, which activates a starch amylase (BAM1) to trigger augmented callose deposition against P. cucumerina [37]. A similar role of ABA and starch metabolism has also been reported in mycorrhiza-induced callose priming in tomato plants upon infection by necrotrophic fungal pathogen B. cinerea [24]. Intriguingly, ABA can enhance or repress Flg22-induced callose deposition in Arabidopsis depending on growth conditions [35] (Figure 1A). Under most growth conditions, ABA has a stimulatory effect on Flg22-induced callose deposition. However, ABA reduced basal and PAMP-induced callose at low light intensity, high sucrose concentration and vitamins (Figure 1A). ABA also represses Flg22-induced callose in hydroponic Arabidopsis grown with the a relatively high concentration of sucrose in the growth medium (0.5%) [34][35] (Figure 1A). These results suggest that ABA’s role in defense-related callose deposition and priming is influenced by carbohydrate metabolism.
3.2. Biogenesis and Activation of Pathogen-Responsive Callose Synthases
As a large transmembrane protein, GSL5/PMR4 belongs to the family of glucan synthases that include plant cellulose synthases. Plant cellulose synthases are synthesized on the endoplasmic reticulum (ER) and go through the ER quality control system for proper folding and modification before being assembled into complexes in the Golgi apparatus and transported to the plasma membrane through the Golgi- and post-Golgi trafficking system [38]. Many proteins involved in plant immune responses, including pattern-recognition receptors and extracellular defense-related proteins, are also synthesized on the ER, go through the ER quality control, and ultimately are transported to their destinations through the secretory pathway. We have recently shown that two Arabidopsis homologs of UBAC2, a conserved ER protein implicated in ER quality control and ER-associated degradation (ERAD), play a critical role in pathogen- and PAMP-induced callose deposition [39]. The UBAC2 proteins interact with the plant-specific PAMP-INDUCED COILED COIL (PICC) protein, which is also localized in the ER and is important for pathogen- and PAMP-induced callose deposition (Figure 2). Therefore, the evolutionarily conserved UBAC2 and plant-specific PICC proteins are critical components of an ER pathway with an important role in plant immunity by regulating pathogen-induced callose deposition. The compromised phenotypes of the ubac2 and picc mutants in PTI and pathogen/PAMP-induced callose deposition could be rescued by overexpression of PMR4, suggesting that disruption of UBAC2 and PICC genes may reduce the accumulation of PMR4 callose synthase in the ubac2 and picc mutants, leading to defects in pathogen/PAMP-induced callose deposition [39]. Further analysis using both GFP- and myc-labeled PMR4 indeed discovered that disruption of UBAC2 or PICC reduced the levels of plasma-membrane-localized PMR4 [39]. These results indicate that the evolutionarily conserved UBAC2 proteins function in coordination with a plant-specific protein PICC in the positive regulation of the biogenesis of PMR4, either by stabilizing the callose synthase or degrading a negative regulator of PMR4 stability (Figure 2).
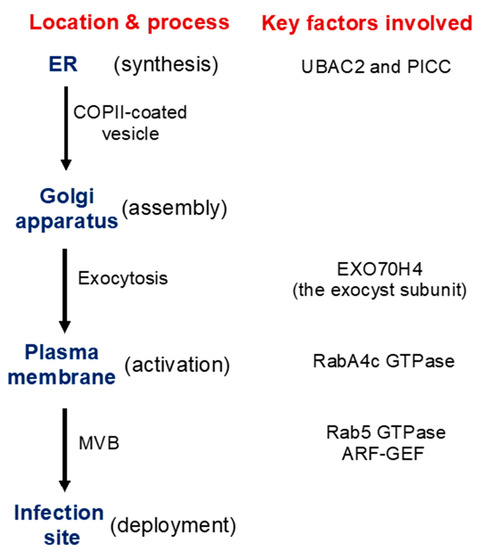
Figure 2. Synthesis, transport, activation, and recruitment of Arabidopsis PMR4 callose synthase during plant defense responses. Some of the important factors identified to be involved in these processes are indicated.
In addition, there is evidence for PMR4 activation in response to pathogen infection or other stress signals. In uninfected PMR4-overexpression lines, no significant increase in the callose synthase activity or callose deposition was observed [40]. In yeast, activation and translocation of callose synthases involve GTPases [41][42]. In Arabidopsis, PMR4 interacts with GTPase RabA4c [43] (Figure 2). Similar to PMR4 overexpression, RabA4c overexpression leads to complete penetration resistance to the virulent powdery mildew fungal pathogen in a PMR4-dependent manner [43]. By contrast, overexpression of a dominant negative form of RabA4c fails to increase callose deposition or penetration resistance [43]. These results indicate that the RabA4c GTPase plays a critical role in the activation and translocation of PMR4 during pathogen-induced callose deposition. As a transmembrane protein synthesized on the ER, PMR4 is likely subjected to additional regulatory mechanisms for its biogenesis, trafficking, and activation to ensure biosynthesis of callose in a timely manner.
3.3. Transport Processes in Pathogen-Induced Callose Deposition
Exocytosis is a process by which cells are able to not only move materials from within a cell to the exterior of the cell, but also insert membrane proteins (such as ion channels and cell surface receptors), lipids, and other components into the cell membrane [44]. Vesicles containing these membrane components fully fuse with and become part of the plasma membrane. The exocyst, an octameric protein complex involved in exocytosis, tethers and spatially target post-Golgi vesicles to the plasma membrane prior to vesicle fusion [44]. It has been demonstrated that the exocyst subunit EXO70H4 is important in the deposition of the callose-rich cell wall layer for biogenesis of the trichome secondary cell wall [45][46]. PMR4 is a callose synthase responsible for the synthesis of callose in the trichome and PMR4 colocalizes with EXO70H4 on plasma membrane microdomains that do not develop in the exo70h4-1 mutant. Significantly, EXO70H4 expression is induced by pathogen elicitor Flg22, which induces callose deposition, in epidermal pavement cells [46]. These results raise a strong possibility that EXO70H4-dependent exocytosis is also involved in the trafficking of newly synthesized PMR4 callose synthase proteins from the Golgi apparatus to the plasma membrane (Figure 2).
Another transport process important for defense-related callose deposition is the recruitment of callose synthases protein such as PMR4 in Arabidopsis from the plasma membrane and transport in vesicle-like bodies to the site of attempted penetration after fungal infection (Figure 1). There is strong evidence that multivesicular bodies (MVBs) are involved in the transportation and delivery of defense components to the forming papilla. Studies have showed accumulation of MVBs and cell wall-associated paramural bodies (PMBs) in the vicinity of pathogen-induced papillae [47][48][49]. PMBs, which are situated between the cell wall and the plasma membrane, are likely to have resulted from the fusion of MVBs with the plasma membrane [50]. Plant MVBs and PMBs have been observed near papillae in plant cells infected by pathogenic fungal, bacteria and nematodes for delivery of defense-related molecules including phytoalexins, callose and ROS to papillae [47][48][51][52][53]. In addition, in plant interaction with filamentous pathogens, each haustorium formed upon successful penetration is surrounded by the plasma membrane of the plant cell termed extrahaustorial membrane (EHM), which is likely synthesized de novo [54]. In tobacco (Nicotiana benthamiana) cells invaded by oomycete pathogen P. infestans, the Rab7 GTPase RabG3c MVB marker protein, but not a tonoplast-localized sucrose transporter, is recruited to the EHM [55]. In Arabidopsis, the Rab5 GTPase, also an MVB marker, accumulates in the EHM after infection with a powdery mildew fungus [56] (Figure 2). Thus, specific rerouting of MVBs from the vacuole to the host-pathogen interface may participate in the formation or modulation of the EHM. In barley, MVBs contained the ADP-ribosylation factor (ARF) GTPase ARFA1b/1c that was important for callose deposition at powdery mildew penetration sites [51]. RNAi knockdown or expression of a dominant negative ARFA1b/1c variant abolished callose accumulation at penetration sites and resulted in increased fungal penetration success. ARFA1b/1c localized to an endosomal MVB compartment that accumulated at fungal penetration sites prior to the accumulation of callose. Interestingly, the Arabidopsis ARF-GEF (guanine nucleotide exchange factor) MIN7 is required for normal levels of callose deposition in response to the Pseudomonas syringae pv. tomato ΔCEL mutant, suggesting that an ARF-dependent vesicle trafficking process may also play a role in callose deposition at sites of pathogen detection in Arabidopsis [57] (Figure 2).
This entry is adapted from the peer-reviewed paper 10.3390/ijms22052393
References
- Stone, B.A.; Clarke, A.C. Chemistry and Biology of (1->3)-b-Glucans; La Trobe University Press: Bundoora, Victoria, 1992.
- Hong, Z.L.; Delauney, A.J.; Verma, D.P.S. A cell plate specific callose synthase and its interaction with phragmoplastin. Plant Cell 2001, 13, 755–768.
- Hong, Z.L.; Zhang, Z.M.; Olson, J.M.; Verma, D.P.S. A novel UDP-glucose transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate. Plant Cell 2001, 13, 769–779.
- Mccormick, S. Male Gametophyte Development. Plant Cell 1993, 5, 1265–1275.
- Barratt, D.H.P.; Kolling, K.; Graf, A.; Pike, M.; Calder, G.; Findlay, K.; Zeeman, S.C.; Smith, A.M. Callose Synthase GSL7 Is Necessary for Normal Phloem Transport and Inflorescence Growth in Arabidopsis. Plant Physiol 2011, 155, 328–341.
- Xie, B.; Wang, X.M.; Zhu, M.S.; Zhang, Z.M.; Hong, Z.L. CalS7 encodes a callose synthase responsible for callose deposition in the phloem. Plant J. 2011, 65, 1–14.
- Wu, S.W.; Kumar, R.; Iswanto, A.B.B.; Kim, J.Y. Callose balancing at plasmodesmata. J. Exp. Bot. 2018, 69, 5325–5339.
- Voigt, C.A. Callose-mediated resistance to pathogenic intruders in plant defense-related papillae. Front. Plant Sci. 2014, 5, 168.
- Osbourn, A.; Carter, J.; Daniels, M.; Dufresne, M.; Hugouvieux, V.; Leggett, M.; Martin-Hernandez, A.; Melton, R.; Morrissey, J.; Papadopoulou, K. Preformed antifungal compounds and plant defence. Biol. Plant-Microbe Interact. 2000, 2, 170–174.
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329.
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351.
- Dou, D.; Zhou, J.M. Phytopathogen effectors subverting host immunity: Different foes, similar battleground. Cell Host Microbe 2012, 12, 484–495.
- Bonardi, V.; Cherkis, K.; Nishimura, M.T.; Dangl, J.L. A new eye on NLR proteins: Focused on clarity or diffused by complexity? Curr. Opin. Immunol. 2012, 24, 41–50.
- Tsuda, K.; Katagiri, F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 2010, 13, 459–465.
- Tsuda, K.; Somssich, I.E. Transcriptional networks in plant immunity. New Phytol. 2015, 206, 932–947.
- Jacobs, A.K.; Lipka, V.; Burton, R.A.; Panstruga, R.; Strizhov, N.; Schulze-Lefert, P.; Fincher, G.B. An Arabidopsis Callose Synthase, GSL5, Is Required for Wound and Papillary Callose Formation. Plant Cell 2003, 15, 2503–2513.
- Nurnberger, T.; Kemmerling, B. PAMP-Triggered Basal Immunity in Plants. Adv. Bot. Res. 2009, 51, 1–38.
- Iriti, M.; Faoro, F. Chemical Diversity and Defence Metabolism: How Plants Cope with Pathogens and Ozone Pollution. Int. J. Mol. Sci. 2009, 10, 3371–3399.
- Hou, S.G.; Liu, Z.Y.; Shen, H.X.; Wu, D.J. Damage-Associated Molecular Pattern-Triggered Immunity in Plants. Front. Plant Sci. 2019, 10.
- Conrath, U. Systemic Acquired Resistance. Plant Signal. Behav. 2006, 1, 179–184.
- Kohler, A.; Schwindling, S.; Conrath, U. Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol. 2002, 128, 1046–1056.
- Ton, J.; Mauch-Mani, B. Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 2004, 38, 119–130.
- Perez-de-Luque, A.; Tille, S.; Johnson, I.; Pascual-Pardo, D.; Ton, J.; Cameron, D.D. The interactive effects of arbuscular mycorrhiza and plant growth-promoting rhizobacteria synergistically enhance host plant defences against pathogens. Sci. Rep. 2017, 7, 16409.
- Sanmartin, N.; Pastor, V.; Pastor-Fernandez, J.; Flors, V.; Pozo, M.J.; Sanchez-Bel, P. Role and mechanisms of callose priming in mycorrhiza-induced resistance. J. Exp. Bot. 2020, 71, 2769–2781.
- Underwood, W. The plant cell wall: A dynamic barrier against pathogen invasion. Front. Plant Sci. 2012, 3, 85.
- Micali, C.O.; Neumann, U.; Grunewald, D.; Panstruga, R.; O’Connell, R. Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell Microbiol. 2011, 13, 210–226.
- Amsbury, S.; Kirk, P.; Benitez-Alfonso, Y. Emerging models on the regulation of intercellular transport by plasmodesmata-associated callose. J. Exp. Bot. 2018, 69, 105–115.
- Lee, J.Y.; Lu, H. Plasmodesmata: The battleground against intruders. Trends Plant Sci. 2011, 16, 201–210.
- Sager, R.E.; Lee, J.Y. Plasmodesmata at a glance. J. Cell Sci. 2018, 131.
- Iglesias, V.A.; Meins, F. Movement of plant viruses is delayed in a beta-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J. 2000, 21, 157–166.
- Galletti, R.; Denoux, C.; Gambetta, S.; Dewdney, J.; Ausubel, F.M.; De Lorenzo, G.; Ferrari, S. The AtrbohD-Mediated Oxidative Burst Elicited by Oligogalacturonides in Arabidopsis Is Dispensable for the Activation of Defense Responses Effective against Botrytis cinerea. Plant Physiol. 2008, 148, 1695–1706.
- Zhang, J.; Shao, F.; Cui, H.; Chen, L.J.; Li, H.T.; Zou, Y.; Long, C.Z.; Lan, L.F.; Chai, J.J.; Chen, S.; et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-Induced immunity in plants. Cell Host Microbe 2007, 1, 175–185.
- Li, Y.; Zhang, Q.Q.; Zhang, J.G.; Wu, L.; Qi, Y.J.; Zhou, J.M. Identification of MicroRNAs Involved in Pathogen-Associated Molecular Pattern-Triggered Plant Innate Immunity. Plant Physiol. 2010, 152, 2222–2231.
- Clay, N.K.; Adio, A.M.; Denoux, C.; Jander, G.; Ausubel, F.M. Glucosinolate Metabolites Required for an Arabidopsis Innate Immune Response. Science 2009, 323, 95–101.
- Luna, E.; Pastor, V.; Robert, J.; Flors, V.; Mauch-Mani, B.; Ton, J. Callose Deposition: A Multifaceted Plant Defense Response. Mol. Plant Microbe Interact. 2011, 24, 183–193.
- Wang, X.; Sager, R.; Cui, W.E.; Zhang, C.; Lu, H.; Lee, J.Y. Salicylic Acid Regulates Plasmodesmata Closure during Innate Immune Responses in Arabidopsis. Plant Cell 2013, 25, 2315–2329.
- Gamir, J.; Pastor, V.; Sanchez-Bel, P.; Agut, B.; Mateu, D.; Garcia-Andrade, J.; Flors, V. Starch degradation, abscisic acid and vesicular trafficking are important elements in callose priming by indole-3-carboxylic acid in response to Plectosphaerella cucumerina infection. Plant J. 2018, 96, 518–531.
- Bashline, L.; Li, S.; Gu, Y. The trafficking of the cellulose synthase complex in higher plants. Ann. Bot. 2014, 114, 1059–1067.
- Wang, Z.; Li, X.; Wang, X.; Liu, N.; Xu, B.; Peng, Q.; Guo, Z.; Fan, B.; Zhu, C.; Chen, Z. Arabidopsis Endoplasmic Reticulum-Localized UBAC2 Proteins Interact with PAMP-INDUCED COILED-COIL to Regulate Pathogen-Induced Callose Deposition and Plant Immunity. Plant Cell 2019, 31, 153–171.
- Ellinger, D.; Naumann, M.; Falter, C.; Zwikowics, C.; Jamrow, T.; Manisseri, C.; Somerville, S.C.; Voigt, C.A. Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol. 2013, 161, 1433–1444.
- Calonge, T.M.; Arellano, M.; Coll, P.M.; Perez, P. Rga5p is a specific Rho1p GTPase-activating protein that regulates cell integrity in Schizosaccharomyces pombe. Mol. Microbiol. 2003, 47, 507–518.
- Qadota, H.; Python, C.P.; Inoue, S.B.; Arisawa, M.; Anraku, Y.; Zheng, Y.; Watanabe, T.; Levin, D.E.; Ohya, Y. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science 1996, 272, 279–281.
- Ellinger, D.; Glockner, A.; Koch, J.; Naumann, M.; Sturtz, V.; Schutt, K.; Manisseri, C.; Somerville, S.C.; Voigt, C.A. Interaction of the Arabidopsis GTPase RabA4c with its effector PMR4 results in complete penetration resistance to powdery mildew. Plant Cell 2014, 26, 3185–3200.
- Liu, J.L.; Guo, W. The exocyst complex in exocytosis and cell migration. Protoplasma 2012, 249, 587–597.
- Kulich, I.; Vojtikova, Z.; Glanc, M.; Ortmannova, J.; Rasmann, S.; Zarsky, V. Cell wall maturation of Arabidopsis trichomes is dependent on exocyst subunit EXO70H4 and involves callose deposition. Plant Physiol. 2015, 168, 120–131.
- Kulich, I.; Vojtikova, Z.; Sabol, P.; Ortmannova, J.; Nedela, V.; Tihlarikova, E.; Zarsky, V. Exocyst Subunit EXO70H4 Has a Specific Role in Callose Synthase Secretion and Silica Accumulation. Plant Physiol. 2018, 176, 2040–2051.
- An, Q.; Ehlers, K.; Kogel, K.H.; van Bel, A.J.; Huckelhoven, R. Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 2006, 172, 563–576.
- An, Q.; Huckelhoven, R.; Kogel, K.H.; van Bel, A.J. Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell. Microbiol. 2006, 8, 1009–1019.
- An, Q.; van Bel, A.J.; Huckelhoven, R. Do plant cells secrete exosomes derived from multivesicular bodies? Plant Signal. Behav. 2007, 2, 4–7.
- Marchant, R.; Robards, A.W. Membrane systems associated with the plasmalemma of plant cells. Ann. Bot. 1968, 32, 457–471.
- Bohlenius, H.; Morch, S.M.; Godfrey, D.; Nielsen, M.E.; Thordal-Christensen, H. The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. Plant Cell 2010, 22, 3831–3844.
- Meyer, D.; Pajonk, S.; Micali, C.; O’Connell, R.; Schulze-Lefert, P. Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. Cell Mol. Biol. 2009, 57, 986–999.
- Nielsen, M.E.; Feechan, A.; Bohlenius, H.; Ueda, T.; Thordal-Christensen, H. Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc. Natl. Acad. Sci. USA 2012, 109, 11443–11448.
- Berkey, R.; Zhang, Y.; Ma, X.; King, H.; Zhang, Q.; Wang, W.; Xiao, S. Homologues of the RPW8 Resistance Protein Are Localized to the Extrahaustorial Membrane that Is Likely Synthesized De Novo. Plant Physiol. 2017, 173, 600–613.
- Bozkurt, T.O.; Belhaj, K.; Dagdas, Y.F.; Chaparro-Garcia, A.; Wu, C.H.; Cano, L.M.; Kamoun, S. Rerouting of plant late endocytic trafficking toward a pathogen interface. Traffic 2015, 16, 204–226.
- Nielsen, M.E.; Jurgens, G.; Thordal-Christensen, H. VPS9a Activates the Rab5 GTPase ARA7 to Confer Distinct Pre- and Postinvasive Plant Innate Immunity. Plant Cell 2017, 29, 1927–1937.
- Nomura, K.; DebRoy, S.; Lee, Y.H.; Pumplin, N.; Jones, J.; He, S.Y. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 2006, 313, 220–223.
