MM is a hematological neoplasm that is still considered an incurable disease. Besides established genetic alterations, recent studies have shown that MM pathogenesis is also characterized by epigenetic aberrations, such as the gain of de novo active chromatin marks in promoter and enhancer regions and extensive DNA hypomethylation of intergenic regions, highlighting the relevance of these non-coding genomic regions. A recent study described how long non-coding RNAs (lncRNAs) correspond to 82% of the MM transcriptome and an increasing number of studies have demonstrated the importance of deregulation of lncRNAs in MM.
- lncRNAs
- multiple myeloma
- RNA-based therapy
1. Introduction
2. Features of lncRNAs
| Gene | Location | Gene Type | Expression in MM | Molecular Mechanism | Molecular Interaction in MM | Biological Effect after lncRNA KD | Biological Effect after lncRNA UR | Prognosis in MM | References |
|---|---|---|---|---|---|---|---|---|---|
| ANRIL | 9p21.3 | Antisense | Up | Decoy | Binds to miR-34a, miR-125a, miR-186 and miR- 411–3p |
Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with worse PFS and OS | [33,34] |
| BM742401 | 18q11.2 | LincRNA | Down | NA | NA | NA | Decreases cell migration | Methylated lncRNA associated with worse OS | [35] |
| Circ_0000190 | 1q42.12 | Circular lncRNA | Down | NA | NA | NA | NA | High expression levels associated with better PFS and OS | [36,37,38] |
| CRNDE | 16q12.2 | LincRNA | Up | Decoy | Binds to miR-451 | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | High expression levels associated with worse OS | [39,40] |
| DARS-AS1 | 2q21.3 | Antisense | Up under hypoxia | Decoy | Interacts with RBM39 and HIP-1α, suppressing mTOR pathway | Decreases cellular proliferation and increases apoptosis. Decreases tumorigenesis in vivo. Its upregulation reduces the sensitivity to bortezomib in vitro | NA | NA | [41] |
| ENSG00000249988 | 4p15.33 | LincRNA | Up | NA | NA | NA | NA | High expression levels associated with worse PFS andbetter OS | [13] |
| ENSG00000254343 | 8q24.12 | LincRNA | Up | NA | NA | NA | NA | High expression levels associated with worse PFS | [13] |
| FEZF1-AS1 | 7q31.32 | Antisense | Up | Decoy | Binds to miR-610 and regulates AKT3 | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | NA | [42] |
| GAS5 | 1q25.1 | Processed transcript | Down | NA | NA | NA | Decreases cellular proliferation | NA | [43] |
| H19 | 11p15.5 | Processed transcript | Up | Decoy | Binds to miR-152-3p and miR-29b-3p | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | High expression levels associated with worse PFS | [44,45,46] |
| HOTAIR | 12q13.31 | Antisense | Up | NA | Activates NF-κB pathway | Decreases cellular proliferation, triggers cell cycle arrest and decreases chemoresistance to dexamethasone | NA | NA | [21,47,48,49] |
| HOXB-AS1 | 17q21.32 | Antisense | Up | Scaffold | Scaffold for ELAVL1. Interacts with FUT4-mediated Wnt/β-catenin pathway | Decreases cellular proliferation and increases apoptosis | NA | NA | [50] |
| IRAIN | 15q26.3 | Antisense | Down | Decoy | Binds to miR-125b and regulates IGF-1 signaling | NA | Increases apoptosis | NA | [51,52] |
| LINC00152 | 2p11.2 | LincRNA | Up | Decoy | Binds to miR-497 | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest. Decreases tumorigenesis in vivo | NA | High expression levels associated with worse OS | [53] |
| LINC00461 | 5q14.3 | LincRNA | Up | NA | NA | Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with worse OS | [54,55,56] |
| LINC00515 | 21q21.3 | LincRNA | Up | Decoy | Binds to miR-140-5p | Increases apoptosis | NA | NA | [57] |
| LINC00665 | 19q13.12 | LincRNA | Up | Decoy | Binds to miR-214-3p | Decreases cellular proliferation and increases apoptosis | NA | NA | [58] |
| LINC01234 | 12q24.13 | LincRNA | Up | Decoy | Binds to miR-124-3p | Decreases cellular proliferation and increases apoptosis. Decreases cell proliferation and tumor growth in vivo | NA | High expression levels associated with worse OS | [59] |
| lnc-ANGPTL1-3 | 1q25.2 | Antisense | Up | Decoy | Binds to miR-30a-3p | Increases the sensitivity to bortezomib | NA | High expression levels associated with worse OS | [60] |
| lnc-TCF7 | 5q31.1 | NA | Up | NA | NA | NA | NA | High expression levels associated with worse PFS and OS | [61] |
| LUCAT1 | 5q14.3 | LincRNA | Up | NA | Activates the TGF-β signaling pathway | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | High expression levels associated with shorter five-year survival | [62] |
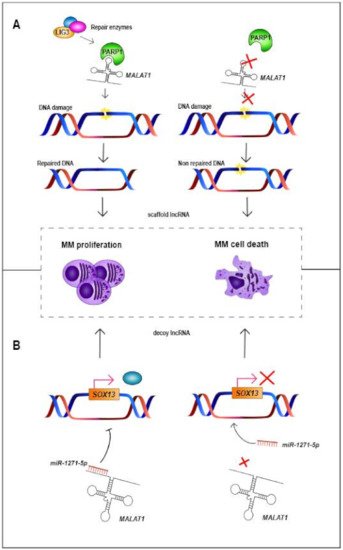
| MALAT1 | 11q13.1 | LincRNA | Up | Decoy and Scaffold | Binds to miR-1271-5p, miR-181a-5p and miR-509- 5p. Scaffold for PARP1 |
Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with worse PFS and OS | [63,64,65,66,67,68,69,70] |
| MEG3 | 14q32.2 | LincRNA | Down | Decoy | Binds to miR-181a | NA | Decreases cellular proliferation and increases apoptosis | High expression levels associated with better PFS and OS | [71,72,73,74] |
| MIAT | 22q12.1 | LincRNA | Up | Decoy | Binds to miR-29b | Sensitizes MM cells to bortezomib | NA | High expression levels associated with worse PFS and OS | [75,76] |
| NEAT1 | 11q13.1 | LincRNA | Up | Decoy | Binds to miR-214 and miR-125a | Decreases cellular proliferation | NA | High expression levels associated with worse PFS and OS | [77,78,79,80] |
| OIP5-AS1 | 15q15.1 | Processed transcript | Down | Decoy | Binds to miR-410 and miR-27a-3p | NA | Decreases cellular proliferation and increases apoptosis | NA | [81,82] |
| PCAT-1 | 8q24.21 | LincRNA | Up | Decoy | Binds to miR-129 | Increases apoptosis and sensitizes MM cells to bortezomib | NA | NA | [83,84] |
| PDIA3P | 1q21.1 | Pseudogene | Up | NA | NA | Decreases cellular proliferation. Increases the sensitivity to bortezomib | NA | High expression levels associated with worse OS | [85] |
| PDLIM1P4 | 3q12.1 | Pseudogene | Up | NA | NA | NA | NA | High expression levels associated with worse PFS and OS | [13] |
| PRAL | 17p13.1 | NA | Down | Decoy | Binds to miR-210 | NA | Decreases cellular proliferation and increases apoptosis. Increases the anti-tumor effect of bortezomib | High expression levels associated with better PFS and OS | [86,87] |
| PVT1 | 8q24.21 | Processed transcript | Up | Decoy | Binds to miR-203a. It is inhibited by BRD4 | Decreases cellular proliferation and increases apoptosis | NA | NA | [88,89] |
| SMILO | 1q42.2 | LincRNA | Up | NA | Regulates IFN pathway | Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with better OS | [13] |
| SNHG16 | 17q25.1 | Processed transcript | Up | Decoy | Binds to miR-342-3p | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | NA | [90] |
| SOX2OT | 3q26.3 | Sense overlapping | Up | Decoy | Binds to miR-144-3p | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest. Decreases tumor growth in vivo | NA | NA | [91] |
| ST3GAL6-AS1 | 3q12.1 | Antisense | Up | NA | NA | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | High expression levels associated with worse PFS | [92,93] |
| TUG1 | 22q12.2 | Antisense | Up | Decoy | Binds to miR-29b-3p and targets HDAC4 | Decreases cellular proliferation and increases apoptosis | NA | NA | [43,94] |
| UCA1 | 19p13.12 | Processed transcript | Up | Decoy | Binds to miR-1271-5p and miR-331-3p | Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with worse OS | [95,96] |
| XLOC_013703 | 20p11.21 | NA | Down | NA | Involved in NF-κB signaling activation | NA | Decreases cellular proliferation and increases apoptosis | NA | [97] |
3. Role of lncRNAs in the Pathobiology of MM
This entry is adapted from the peer-reviewed paper 10.3390/cancers13081976
