Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biology
The fruit of Lycium barbarum L. (goji berry) is used as traditional Chinese medicine, and has the functions of immune regulation, anti-tumor, neuroprotection, anti-diabetes, and anti-fatigue. One of the main bioactive components is L. barbarum polysaccharide (LBP).
- lycium barbarum polysaccharide
1. Introduction
There are more than 80 Lycium species in the world, and they are widely distributed in Asia, Europe, North America, and other regions [1]. Among them, Lycium barbarum L. is a perennial deciduous shrub, and its fruits, goji berries, have been used as one of the traditional Chinese drugs [2,3]. Currently, goji berry is recorded in the Pharmacopoeia of the People’s Republic of China [4,5]. Goji berry has nourishing effects on the kidneys and lungs [6]. It is regarded as a nourishing Chinese medicine, and can be used as medicine and food [7]. Goji berry has diverse biological activities, including anti-inflammatory [8], anti-tumor [9], anti-oxidation [7], anti-aging [10], hypoglycemic [11], and hypolipidemic [12].
Goji berry is rich in nutrition and abundant in natural products, such as L. barbarum polysaccharide (LBP), betaine, flavone, and vitamin [3]. Among its bioactive components, LBP composes 5%–8% of the dried goji berry [13]. LBP is a water-soluble complex with carbohydrate chains and proteins; the carbohydrate chains are mainly composed of six saccharides, including arabinose, galactose, glucose, rhamnose, mannose, and xylose, which accounts for about 70% of all the saccharides [14,15]. LBP can modulate gut microbiota to improve nutrient utilization and health [16,17]. Nowadays, LBP is mainly extracted from goji berry. It takes one year to obtain goji berry, and climate, diseases as well as insect pests might affect the yield of goji berry [18]. Moreover, planting areas for high-quality goji berry with high-levels of LBP are limited. Besides, fresh goji berry is highly perishable, which limits the acquisition of high-quality LBP [19,20]. Extraction of LBP from goji berry is unable to meet the rapidly increasing commercial LBP demands. Therefore, it is of great interest to find other sustainable and stable LBP supplies [7].
With the rapid development of yeast synthetic biology, Saccharomyces cerevisiae and other yeasts have been used as cell factories for the production of plant natural products [21]. One advantage of producing polysaccharides using yeast cell factories is that the production is not affected by seasons, regions, and pests [22]. S. cerevisiae is generally recognized as a safe microorganism (GRAS) and has been widely used in food and drug production. Some plant natural products have been produced in engineered S. cerevisiae, such as cocoa butter and ginsenosides [23,24,25,26,27,28,29,30,31]. Therefore, S. cerevisiae is an ideal microbial host to produce LBP. Introducing the LBP biosynthetic pathway and rewiring the metabolic pathway of S. cerevisiae might provide a green and feasible way for LBP production [32,33].
2. LBP Biosynthetic Pathway in L. barbarum
LBP is constituted of six saccharides of α-(1→4)-GalA, α-(1→6)-Glc, β-(1→3)-Galp, β-(1→6)-Galp, α-(1→5) -Ara, and β-(1→4)–Galp. Enzymatic selectivity is essential to combine the diverse saccharides into LBP. After the glycosidic bond is formed, the stereochemistry of LBP can be retained or reversed by glycosyltransferases [34,35,36]. In fact, multiple metabolic pathways might participate in LBP biosynthesis in goji berry, including the galactose metabolism pathway, tricarboxylic acid cycle, glyoxylic acid cycle, propionic acid metabolism, amino sugars metabolism, starch metabolism, and sucrose metabolism. These pathways provide precursors for the anabolism of various carbohydrates [37,38]. Among these pathways, sucrose synthetic pathway is mainly responsible for plant cell wall formation and biomass accumulation. Sucrose phosphate synthase (SPS) and sucrose phosphate phosphatase (SPP) are the two essential enzymes in sucrose biosynthesis [39]. In addition, the acid invertase (AI), sucrose synthase (SS), and SPS involved in sucrose metabolism are the key enzymes of the plant carbon metabolic pathway [40]. Sucrose synthesis is closely associated with the metabolism of amino sugar, nucleotide sugar, and galactose [41]. The fruits of goji berry mainly synthesize glucose and fructose, and the LBP content is mainly determined by fructose content in goji fruits.
The α-galactosidase (GALA) participates in several steps of galactose metabolism. These carbohydrate anabolic pathways are the prerequisite for LBP synthesis, and they can regulate carbohydrate accumulation. UDP-glucose pyrophosphorylase (UGP) is an essential enzyme involved in carbohydrate metabolism, which can affect normal cell development, polysaccharide synthesis, and stress response in S. cerevisiae and other fungi [42,43]. The key enzymes in the LBP synthetic pathway, UDP-glucuronate 4-epimerase (GAE), malate synthase (MS), and α-galactosidase (GALA), have not been characterized yet [37]. These key genes play important roles in the synthesis of LBP, and the information might be obtained by analyzing the transcriptome of goji berry (Figure 1).
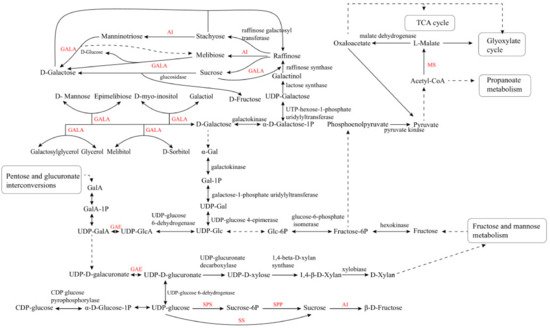
Figure 1. Metabolic pathways of saccharides in goji berry. AI, acid invertase; GAE, UDP-glucuronate 4-epimerase; GALA, α-galactosidase; MS, malate synthase; SPS, sucrose phosphate synthase; SPP, sucrose phosphate phosphatase; SS, sucrose synthase.
3. Mining of Key Enzymes for LBP Biosynthesis
The transcriptome of goji berry can help recover genes in the LBP biosynthetic pathway. Several studies about transcriptome of goji berry have been applied. Wang et al. prepared the leaf transcriptome of goji berry and elucidated the mechanisms of carotenoid biosynthesis in goji berry [44]. Chen et al. obtained 139,333 predicted genes of goji berry, and they identified genes in the flavonoid and taurine biosynthetic pathways [45]. Ma et al. analyzed the transcriptome of goji berry and identified the candidate genes involved in sugar metabolism under elevated CO2, which helped recover the LBP biosynthetic pathway [37]. However, current omics data in public databases have not recovered the whole LBP biosynthetic pathway in goji berry.
To explore the key enzymes in the LBP biosynthetic pathway, collecting fresh tissues of goji berry at different growth phases and different planting areas are necessary. Sequencing of these fresh tissues would give an array of transcriptome data, and de novo assembly of these data would obtain full gene profiles of goji berry. Further transcriptome comparison analysis would give insights into the details of the LBP biosynthetic pathway (Figure 2). To predict the key enzymes in LBP synthetic pathways, phylogenetic, gene similarity network analysis, and other bioinformatic methods can be used to identify the candidate key genes [46]. Diverse glycosyltransferase genes were identified from public omics data by using phylogenetic analyses and plant secondary product glycosyltransferase (PSPG) homolog [28,29,30]. Thus, finding homologous in goji genomes is possible to predict potential glycosyltransferases for LBP synthesis. As the sugar metabolic pathway is complex, characterizing candidate LBP synthetic genes identified with bioinformatic strategies will help identify efficient enzymes for LBP biosynthesis in engineered yeasts.
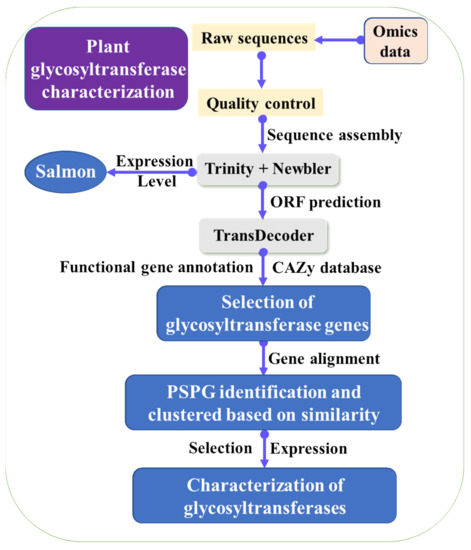
Figure 2. Strategy used for identification and characterization of glycosyltransferase genes from transcriptomic data.
The glycosyltransferases can determine saccharide compositions and arrangements of the LBP. The bioinformatic strategy was used for the recovery of glycosyltransferases functioned in ginsenoside biosynthesis. The omics data in the public database were downloaded and assembled. The open reading frames (ORFs) were predicted and annotated with the help of CAZy and other databases. The candidate glycosyltransferase genes were selected for sequence alignment. Only predicted glycosyltransferase genes with more than 1320 bp and PSPG motif were used for further analysis. The glycosyltransferase genes were clustered based on the similarity, in order to reduce the redundancy. Besides, gene expression level can be determined at different growth stages, which can help identify real glycosyltransferase genes. The identified potential glycosyltransferase genes for ginsenosides were selected for expression. The enzymatic characterization of the glycosyltransferases was identified (Figure 2), and the characterized glycosyltransferases were used for adding activated sugars to the substrates [28,29,30,31]. Recently, another two cellulose synthases were identified to function as glycosyltransferases in saponin biosynthesis [47,48], showing identification of glycosyltransferases for LBP synthesis is complex. Nowadays, the use of gene similarity and network analysis have been widely used for gene identification [46].
This entry is adapted from the peer-reviewed paper 10.3390/molecules26061641
This entry is offline, you can click here to edit this entry!
