Aluminum (Al) toxicity is a major environmental stress that inhibits plant growth and development in acidic soils.
- Al toxicity
- Al tolerance mechanism
- Al tolerance strategy
1. Introduction
Aluminum (Al) is the third most abundant chemical element in the earth’s crust. Al mainly exists as aluminosilicates and oxides with non-phytotoxicity in neutral or slightly acidic soils. In acidic conditions (pH < 5.0), the mineral form of Al dissolves to release the soluble Al3+ species, which can rapidly inhibit root elongation and further affect the uptake of water and nutrients, eventually resulting in nutritional deficiency and drought stress, which lead to severe loss of plant productivity [1][2]. Acidic soils occupy approximately 50% of potentially arable lands worldwide, most of which are distributed in Southwest Asia, Central Africa, and South America, as well as in Australia, eastern North America, and throughout Europe [3][4]. In recent decades, with an increasing intensity of human activities, especially an increase in acid deposition caused by global industrialization and the high-intensity utilization of agricultural soil have led to the continuous entry of a large amount of exogenous H+ into the soil, which has greatly accelerated the process of soil acidification, raising a huge threat to sustainable agricultural development and food security [5]. Liming can ameliorate Al toxicity by raising the soil pH, but it has little effect on the improvement of the underlying soil and hardens the soil structure [6]. The world’s population is also growing rapidly and is anticipated to increase to 9.7 billion by 2050 [7]. Therefore, understanding Al-resistance mechanisms and the development of strategies to confer plant resistance for sustainable agricultural productivity remains imperative.
The research on Al stress has been updated rapidly in recent years, and a substantial number of new genes have been proven to be involved. For example, Snowden et al. cloned five wali1–5 (wheat aluminum induced) genes from the root tips of Al-treated Warigal wheat (Triticum aestivum) [8]. Subsequently, Richards et al. cloned wali6 and wali7 in this variety of wheat [9]. However, there are limited reviews on Al stress. Furthermore, more and more evidence has confirmed the involvement and roles of Al in promoting plant growth, improving phosphorus efficiency, and alleviating H+, manganese, and iron toxicities in acidic conditions [10][11]. Additionally, Al stress confers plants tolerance to abiotic stresses by activating the stress-related genes and attracts the plant growth-promoting rhizobacteria (PGPRs) toward roots by inducing root exudates [10][12]. Therefore, more reviews on the progress of research on Al stress are needed.
2. Effects of Aluminum (Al) Stress in Plants
Al stress has become the main limiting factor with multifarious detrimental effects in plants (Figure 1). The Al3+ ion is a multivalent cation that rapidly and strongly binds to negatively charged sites in the root [13]. It has been reported that Al changes the properties of the cell wall (CW) and interferes with the transport of molecules across the cell membrane, influencing an array of intercellular processes [14]. The major target site of Al toxicity is the root apex, particularly the distal part of the transition zone [15]. Al binding of the root causes loss of Mg2+, K+, and Ca2+, as well as limits the availability of indispensable nutrients, especially phosphorus (P), magnesium (Mg), and molybdenum (Mo), therefore, impairing root growth [16][17]. Al also reduces expansion, stomatal closure, and net photosynthesis in leaves, and tends to bind with P to form insoluble complexes in acidic soil, thereby resulting in P deficiency for plant growth [18][19]. Likewise, Al stress decreases the phytoextraction capability from contaminated soils using hyperaccumulators [20]. Moreover, Al stress can cause the production of reactive oxygen species (ROS) and reactive nitrogen species (RNS), which trigger a series of free radical chain reactions, including the peroxidation of the cell membrane system, decrease in enzyme activities, decomposition of chlorophyll, and breakage of the DNA strand [21]. Recently, Al stress has been reported to disturb soil rhizobia by affecting the efficacy of nodulation and N-fixation in legume species, and therefore influences the balance of hormones in plant roots, which has been proposed to cause growth inhibition [22][23]. Intriguingly, Al3+ has been regarded as a beneficial element in the growth of some plants in acidic soil, and Al-induced growth enhancement in tea plants has been associated with the maintenance of DNA integrity in meristematic cells and increased uptake of nutrient elements [10][24][25].
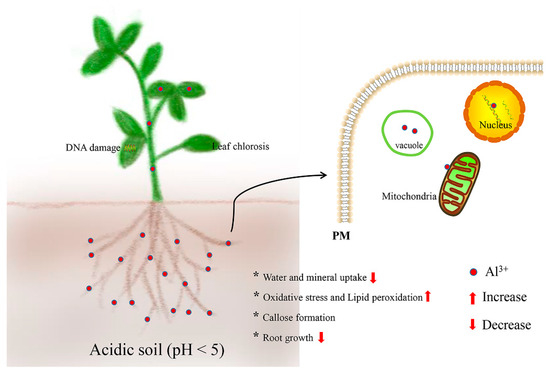
Figure 1. Adverse detrimental effects of aluminum (Al) stress and their location in plants.
3. Al Tolerance Mechanisms in Plants
Plants thriving in acidic soil have adaptations, including external exclusion and internal tolerance, to detoxify Al [3][26]. External exclusion prevents Al from entering cells through Al-induced root exudation of organic compounds into the rhizosphere, decreasing the capacity of CWs to bind Al3+ and rhizosphere alkalization. Internal tolerance mainly includes the uptake and sequestration of Al in the vacuole and the improvement of antioxidant capacity inside cells [5][27]. Recently, various approaches have been used to study the mechanisms of Al tolerance in plants. Transcriptome, proteome, metabolome, mutation breeding methodologies, and several Al-tolerant genes have been reported to be involved in the secretion of organic acids (OAs) for Al3+ chelation, CW modification for the reduction in Al content, pH increase in the rhizosphere, etc. (Table 1).
Table 1. Genes confirmed to be involved in Al tolerance in recent years.
| Genes | Description | Plant Species | Functions | References |
|---|---|---|---|---|
| AhFRDL1 | Ferric reductase defective-like 1 protein | Arachis hypogaea | Transport citrate | [28] |
| AtPrx64 | Class III peroxidase | Arabidopsis thaliana | Peroxidase | [29] |
| AvSAMS1 | S-adenosyl methionine synthetase | Andropogon virginicus | Alterations of methylation status | [30] |
| BoALMT1 | Aluminum induced malate transporter | Brassica oleracea | Transport malate | [31] |
| BdMATE | Multidrug and toxic compound extrusion | Brachypodium distachyon | Transport citrate | [32] |
| GmMATE75 | Glycine max | [33] | ||
| GmMATE79 | Glycine max | [33] | ||
| GmMATE87 | Glycine max | [33] | ||
| GsMATE | Glycine soja | [34] | ||
| PtrMATE1 | Populus tomentosa | [35] | ||
| PtrMATE2 | Populus tomentosa | [35] | ||
| TaMATE1B | Triticum aestivum | [36] | ||
| FeSTAR1 | Half-type ABC transporter | Fagopyrum esculentum | Affect cell wall hemicellulose metabolism | [37] |
| GmGRPL | Glycine-rich protein-like protein | Glycine max | Regulating the level of indole-3-acetic acid (IAA) and ethylene | [38] |
| GmIREG3 | Iron regulated/ferroportin | Glycine max | Sequestrating Al into the vacuoles | [39] |
| GmME1 | NADP-malic enzyme | Glycine max | NADP-malic enzyme activity | [40] |
| HtNHX1 | Sodium (potassium)/proton antiporters | Helianthus tuberosus | Na+/H+ antiporter | [41] |
| HtNHX2 | Helianthus tuberosus | [41] | ||
| HvABCB25 | ATP binding cassette transporters | Hordeum vulgare | Vacuolar Al sequestration | [42] |
| MsCS | Citrate synthase | Medicago sativa | Citrate synthesis | [43] |
| MsPG | Polygalacturonase | Medicago sativa | Decreasing Al accumulation and increasing porosity and extensibility of cell walls | [44] |
| NtSUT1 | Sucrose transporter | Nicotiana tabacum | Sucrose uptake | [45] |
| OsAUX3 | Auxin carrier | Oryza sativa | Auxin influx carrier | [46] |
| VuAAE3 | Acyl activating enzyme | Vigna umbellata | Oxalyl-CoA synthetase | [47] |
| VuFDH | Formate dehydrogenase | Vigna umbellata | Catalyze the oxidation of formate | [48] |
| ZjOMT | Methyltransferase | Zoysia japonica | Melatonin synthesis | [49] |
| ZmAT6 | Aluminum tolerance protein | Zea mays | Scavenging reactive oxygen species | [50] |
| ZmPGP1 | P-glycoprotein | Zea mays | Auxin efflux carrier | [51] |
3.1. External Exclusion Mechanisms
In the Al tolerance mechanism, plants can secrete OAs, such as citrate, malate, and oxalate, from roots under Al stress [3][26][52]. Accumulating evidence has shown that citrate is more dominant than malate and oxalate in response to Al stress [3]. Al-induced secretion of OAs is mediated through anion channels or transporters [53]. To date, Al-activated malate transporter (ALMT) and multidrug and toxic compound extrusion (MATE) have been identified [5][16][17][27]. Since Sasaki et al. identified TaALMT1 from wheat [54], it has been discovered that ALMT is involved in Al-induced secretion of malate to alleviate Al toxicity in Arabidopsis [55][56], barley (Hordeum vulgare) [57], rape (Brassica napus) [58][59], maize (Zea mays) [60], Yorkshire fog (Holcus lanatus) [61], Camelina (Camelina sativa) [62], rubber tree (Hevea brasiliensis) [63], and cabbage (Brassica oleracea) [31], etc. Interestingly, TaALMT1 also plays a role in alkaline tolerance by promoting exudation of both malate and gamma-aminobutyric acid (GABA) in wheat. In addition, MATEs, which are well characterized as multidrug transporters, can transport various substrates, such as citrate, secondary metabolites, and plant hormones, through electrochemical cation gradients [64]. In recent years, MATE genes have been identified from various plant species, including Arabidopsis [65], cabbage [66], Brachypodium distachyon [32], eucalyptus (Eucalyptus camaldulensis) [67], buckwheat (Fagopyrum esculentum) [68], soybean (Glycine max) [33][69][70], Rubiaceae (Psychotria rubra) [71], sorghum (Sorghum bicolor) [72], rice bean (Vigna umbellata) [73][74][75][76], maize [77][78], wheat [36], wild soybean (Glycine soja) [34], etc. The difference in cell location and Al-induced expression patterns of MATE transporters have been determined. For instance, FeMATE1 localized on the plasma membrane (PM), is specifically expressed in roots, whereas FeMATE2 located on the Golgi membrane, is expressed in both the roots and the leaves [68]. Al stress could induce PtrMATE1 expression in 12 h, in contrast to the expression pattern of PtrMATE2, which occurred 24 h after Al3+ treatment [35]. VuMATE1 was expressed at 6 h after Al stress, whereas VuMATE2 was expressed at the early stage of Al stress [73][76]. However, most plants that adopt external exclusion need to protect the root tip from Al toxicity directly on the root surface and ensure the dynamic balance of plant essential nutrients [79]. Al is the most abundant metal element in the earth’s crust; plants cannot alleviate all Al toxicity in acidic soil but can neutralize a portion around the root tip. Therefore, the first reaction of plants to Al toxicity is to prevent the Al3+ around the root tip from entering the root cells, which may be the main target of OAs [80].
The transmembrane transport of OAs depends on the driving force formed by the proton electrochemical gradient on both sides of the PM, which provides continuous power for organic acid secretion [81][82]. Al stress increases the activity of PM H+-ATPase and inhibits the expression of VHA-a2, VHA-a3, and vacuolar proton pump activity, and therefore increases the proton gradient on both sides of the PM and promotes OAs secretion by AtALMT1 and AtMATE [83]. Al stress reduces the entry of OAs into vacuoles by inhibiting vacuolar proton pump activity and activating the PM transport system, which secretes large amounts of intracellular OAs to the apoplast. While OAs secretion is blocked, vacuolar proton pump activity is activated, indicating that intracellular Al tolerance may be an alternative [83]. Additionally, it is known that OAs secretion depends on the maintenance of internal concentrations of OAs. Zhou et al. identified a cytosolic NADP-malic enzyme, GmME1, which was implicated in the organic acid pool and confers higher Al resistance by increasing internal malate and citrate concentrations and their external efflux [40]. Recently, Sun et al. found that the level of the MsCS transcript was higher in Al tolerant cultivar as compared with the Al sensitive cultivar and the activity of citrate synthase (CS) affected Al resistance through citrate concentration and exudation in alfalfa cultivar [43]. These results suggest that OAs secretion can be regulated through the activation of OA transporters, PM H+-ATPase activity, and improvement of the internal OAs pool.
In addition to the secretion of OAs, the release of phenols has been verified to have the capacity to detoxify Al through chelating Al3+ ions. For example, Al and silicon (Si) trigger the release of catechol, catechins, and quercetin by root tips, which could potentially detoxify Al [84]. Ma et al. also found that phenolic compounds were involved in coping with Al toxicity in the Chinese fir by comparative transcriptome [85]. Chen et al. reported the enhancement of polyphenolic metabolism as an adaptive response to Al stress in lettuce roots [86]. Recently, the findings of Fu et al. indicated that Al-polyphenol complexes improved Al resistance in tea plants [87]. Therefore, a comprehensive exploration of the roles of phenolic compounds under Al stress might be an ideal target for genetic engineering in the future.
The CW is the first barrier for plants to resist external stress and has been recognized as the major target of Al toxicity; plants can alleviate Al stress by modifying the CW [88]. For example, OsSTAR1 interacts with OsSTAR2 to form an ATP-binding cassette (ABC) transporter complex, which is specifically responsible for transporting uridine diphosphate (UDP)-glucose and modifying CWs in rice [89]. Similarly, FeSTAR1 and FeSTAR2 also form an ABC transporter complex, which participates in Al tolerance through the CW matrix polysaccharide metabolism in buckwheat [37][90]. Recently, Fan et al. showed that abscisic acid (ABA) alleviation of Al toxicity in rice beans depends on ABI5-mediated CW modification and osmoregulation [91]. More recently, Liu et al. identified a 4-coumarate, which influences Al resistance through the modification the CW [46]. Moreover, pectin methylesterase (PME) plays an important role in Al tolerance by regulating the degree of pectin esterification in the CW, which converts highly methylated pectin into a negatively charged demethylation form, resulting in more Al3+ binding to pectin [92]. Overexpression of OsPME14 accumulated the content of Al in the root tip CW and increased its sensitivity to Al in rice [93]. Furthermore, it was found that Al-tolerant varieties showed higher methylated pectin ratio and lower PME activity in rice, corn, and buckwheat [13][94][95]. Therefore, Al-resistant crop varieties can be cultivated by reducing the pectin content of plant root tips or increasing the degree of methylation of pectin.
Additionally, rhizosphere alkalization is one of Al tolerance mechanisms by reducing the solubility of Al [96]. For example, Yang et al. showed that elevated pH from 2.5 to 4.0, alleviates the Al-toxicity of Citrus by increasing root accumulation of malate and citrate [97][98]. Further experiment showed that raised pH alleviates Al-induced a decrease in total soluble protein level and an increase in electrolyte leakage in citrus leaves and roots by conferring the ability to maintain a balance between production and detoxification of ROS and methylglyoxal [92]. Moreover, Liu et al.’s study on wild barley showed that low pH and higher Al tolerance in XZ16 were associated with a higher ability of H+ uptake and rhizospheric alkalization [99]. Magnesium promotes the elevated root surface pH regulation in Populus, resulting in root surface alkalization in the transition zone, which alleviates the toxicity of high concentration Al [100].
3.2. Internal Tolerance Mechanisms
Internal tolerance mechanisms are mainly involved in Al detoxification, uptake, translocation, and accumulation of non-phytotoxic complexes of Al in the various organs [101][102]. For instance, buckwheat, hydrangea, and tea can accumulate a large amount of Al in the aboveground parts via transporting nontoxic Al complexes [103][104][105]. Several transporters have been reported to be involved in the absorption, sequestration, and transportation of Al from roots to aboveground parts in plants. In rice, OsNrat1, a PM-located transporter, belongs to the natural resistance-associated macrophage protein (Nramp) and has low similarity with other Nramp members, which specifically transports Al3+ rather than bivalent metals (Mn2+, Fe2+, and Cd2+) [102]. Bioinformatics suggest that the Ala-Ile-Ile-Thr element is the key determinant of Nrat1 for Al selectivity [106]. In Arabidopsis, AtALS3 acts as an Al transporter to redistribute Al outside sensitive tissues [107]. Moreover, OsALS1, an ABC transporter located in the tonoplast, sequestrates Al3+ in the vacuole. The knockout of OsALS1 leads the high sensitivity of rice to Al stress [26]. FeALS1.1 and FeALS1.2, OsALS1 homologs, can also sequestrate Al3+ in the vacuoles and detoxify Al in the roots and leaves in buckwheat [108]. Likewise, the AvABCG1 transporter confers Al tolerance by accumulating Al3+ in specific areas of Andropogon virginicus [109]. HvABCB25 transports Al from the cytoplasm to the vacuoles for sequestration in barley [42]. In addition, HmPALT1 and HmVALT1 are involved in transporting Al in hydrangea (Hydrangea macrophylla) [105]. The mechanism of Al detoxification in plants is achieved by forming non-phytotoxic complexes of small molecular organic compounds with Al3+ [10][27][52]. NIP1;2, the closest homolog to HmPALT1, facilitates the transport of Al-malate from the CW to the symplast in Arabidopsis [110]. NIP1;2-mediated transport of Al-malate complex depends on Al-induced malate secretion mediated by AtALMT1. Therefore, the coordinated operation of the Al detoxification mechanism between external and internal parts of the plants is linked by NIP1;2 and AtALMT1.
Studies have showed that the transportation of other ions could also alleviate Al toxicity [19]. For example, Ca2+ serves as an essential second messenger to modulate developmental plasticity in plants, which reduces the concentration of active Al and fixation of P under Al stress [111][112]. Treatment with higher Ca2+ concentration alleviates Al-induced inhibition of root growth, which is attributed to higher cytosolic Ca2+ concentrations through specific Ca2+ signatures triggering downstream responses [113]. Moreover, Mg2+ is involved in metabolism-activating enzymes such as CS and malate synthase by functioning as a cofactor for enzymes, thereby activating OA synthesis to alleviate Al toxicity [114]. Overexpression of AtMGT1 and OsMGT1 confers Al tolerance in plants by increasing the absorption of Mg and inhibiting potential targets of Al [115]. Li et al. showed that Mg promoted root growth and increased Al tolerance by modulating the production of nitric oxide in Arabidopsis [116]. Similarly, Kong et al. showed that the addition of Mg to the Al treatment solution alleviated Al-induced inhibition of root growth, suppressed Al uptake, and reduced hydrogen peroxide (H2O2) concentration in maize [99]. Furthermore, K+ efflux was related to Al tolerance by accompanying OA secretion [99]. Recently, Li et al. showed that ectopic expression of either HtNHX1 or HtNHX2, from Jerusalem artichoke (Helianthus tuberosus), could enhance rice tolerance to Al stress and soil acidity by altering K+ and H+ fluxes and the CW structure [41]. In addition, the application of zinc has been shown to alleviate Al-induced damage via competing with Al and increasing the IAA content in alfalfa [46]. Sulfate supplementation activates short-term tolerance to Al toxicity in perennial ryegrass (Lolium perenne) roots by upregulating total superoxide dismutase (SOD) activity [117]. Further studies are required to investigate how the adjustment of the formula of fertilizer may alleviate Al toxicity.
Analogous to other abiotic stimuli, Al stress induces the overproduction of ROS and lipid peroxidation, resulting in serious cell damage and even cell death [118]. To protect plants from Al-triggered oxidative stress, plant tolerance to Al toxicity is enhanced by improving the activity of ROS-scavenging enzymes, reducing the production of ROS, and weakening lipid peroxidation [119]. For example, overexpression of WMnSOD1, an Al-induced SOD, increases oxidative resistance, and Al tolerance [120]. Overexpression of AtBCB and NtGDI1 ameliorates oxidative stress and confers a degree of resistance to Al stress [121]. Overexpression of AtPrx64 reduces the accumulation of ROS and Al, thereby promoting root growth [29]. Recently, ZmAT6, a chloroplast-located protein, has been shown to increase the expression level of the ZmSOD gene and improve the activity of antioxidant enzymes SOD in the antioxidant enzymatic system. In addition, the overexpression of ZmAT6 in maize and Arabidopsis increased the activity of several enzymes within the antioxidant system, thereby enhancing Al toxicity tolerance [50]. Moreover, methyltransferase could reduce ROS, lipid peroxidation, and ion leakage, and overexpression of the methyltransferase gene can improve stress resistance of plants [122]. For example, overexpression of ZjOMT enhanced Al tolerance of Escherichia coli by increasing the content of melatonin [49]. In addition, a metabolic change is an internal tolerance mechanistic in response to Al stress [48]. Overexpression of VuFDH increased Al tolerance, which is likely due to their decreased Al-induced formate production in tobacco (Nicotiana tabacum) [48]. Likewise, VuAAE3 played a critical role in Al tolerance mechanisms via function as oxalyl-CoA synthetase [47].
4. Transcription Factors Are Involved in Adaptation to Al Stress
Transcription factors are protein complexes that regulate the transcription of genetic information from DNA to mRNA via specific binding to cis-acting elements in the promoters of target genes and acting downstream of signaling cascades in response to environmental stress [123]. The role of TFs in the Al signaling pathway has attracted significant attention since the first TF, sensitive to proton rhizotoxicity 1 (STOP1) and involved in Al tolerance, was identified. More TFs have also been identified to be involved in Al-induced signaling pathways (Table 2).
Table 2. Transcription factors involved in Al tolerance.
| TFs | Categories | Plant Species | Functions | References |
|---|---|---|---|---|
| AtHB7 | HD-Zip I transcription factor | Arabidopsis thaliana | Regulate the capacity of the cell wall to bind Al | [124] |
| AtHB12 | Arabidopsis thaliana | [124] | ||
| AtWRKY47 | WRKY transcription factor | Arabidopsis thaliana | Regulating genes responsible for cell wall modification | [125] |
| OsWRKY22 | Oryza sativa | Activation of OsFRDL4 expression and enhancement of citrate secretion |
[126] | |
| CcSTOP1 | C2H2-type zinc finger transcription factor | Cajanus cajan | Regulate genes for OA transporters Regulate GhMATE and GhALMT1 expression Regulate the downstream Al or low pH resistance genes |
[127] |
| GhSTOP1 | Gossypium hirsutum L. | [128] | ||
| GmSTOP1a | Glycine max | [129] | ||
| GsGIS3 | Glycine soja | Regulating Al-tolerance-related genes | [130] | |
| HvATF1 | Hordeum vulgare L. | Regulating multiple downstream genes involved in Al resistance |
[131] | |
| NtSTOP1 | Nicotiana tabacum | Activation of NtMATE expression | [132] | |
| OsART2 | Oryza sativa | Regulate at least four genes implicated in Al tolerance | [133] | |
| SbSTOP1 | Sorghum bicolor L. | Regulate SbMATE and SbSTAR2 expression | [134] | |
| GsMAS1 | MADS-box transcription factor | Glycine Soja | Accumulation of Al-activated citrate and malate |
[135] |
| HvHOX9 | Homeobox-leucine zipper transcription factor | Hordeum vulgare L. | Regulate the capacity of the cell wall to bind Al | [136] |
| MdMYC2 | bHLH transcription factor | Malus domestica | Activation of ethylene biosynthesis | [137] |
| VuABI5 | Basic-leucine zipper transcription factor | Vigna umbellata | Regulate genes involved cell wall modification and osmoregulation | [91] |
| VuNAR1 | NAC-type transcription factor | Vigna umbellata | Regulate cell wall pectin metabolism | [138] |
AtSTOP1, a C2H2-type zinc finger transcription factor, has been found to be critical for both proton and Al tolerance, which regulates the expression of downstream-STOP1 Al-resistance genes. However, the expression of AtSTOP1 is unaffected by Al stress, which suggests that AtSTOP1 is modulated by Al at posttranscriptional or posttranslational levels. Zhang et al. showed that an F-box protein-encoding gene regulation of the Al-activated malate transporter expression 1 (RAE1) regulates the stability of STOP1 via the ubiquitin-26S proteasome pathway in Arabidopsis. This indicates that STOP1 is regulated at a posttranslational level [139]. Recently, Guo et al. showed that hyperrecombination protein 1 (HPR1) regulates nucleocytoplasmic STOP1 mRNA export to modulate the expression of STOP1 downstream genes and Al resistance of plants, highlighting that the regulation of STOP1 by HPR1 occurs at a posttranscriptional level [140]. More recently, Fang et al. showed that the SUMOylation of STOP1 is involved in the regulation of Al resistance [141]. In this study, STOP1 is mono-SUMOylated at K40, K212, or K395 sites; and blocking STOP1 SUMOylation reduced Al resistance through the reduction in STOP1 stability and the expression of STOP1-regulated genes. Moreover, the SUMO protease ESD4 specifically interacts with deSUMOylates STOP1, and mutation of ESD4 increases the SUMOylation of STOP1 and the expression of AtALMT1, which contribute to Al stress tolerance.
The functions of STOP1-like proteins, including CcSTOP1 [127], GhSTOP1 [128], GmSTOP1 [129], NtSTOP1 [132], and SbSTOP1 [134], in other plant species, have been characterized, and proven to be essential for the expression of several Al-tolerance-related genes. For example, GmSTOP1 contributes to both Al resistance and H+ tolerance, and overexpression of GmSTOP1a increases the expression of GmALMT1 and decreases Al accumulation in soybean hairy roots under Al stress [129]. AtSTOP1 and OsART1 are both central regulators involved in Al tolerance through the regulation of multiple downstream genes. However, the rice homolog (Al resistance transcription factor, OsART1) regulates only Al tolerance genes [142]. Furthermore, OsART2, a homolog of OsART1, has been shown to regulate Al tolerance independent of the OsART1-regulated pathway in rice and to play a supplementary role in Al tolerance [133]. HvATF1 (Al-tolerant transcription factor 1) is the closest homolog of AtSTOP1 and OsART1 and alleviates Al stress through regulating multiple genes in barley; this provides insights into the different molecular mechanisms of Al tolerance in plants [131]. Liu et al. also cloned a C2H2 zinc-finger protein, GsGIS3, which enhanced tolerance to Al toxicity by regulating Al-tolerance-related genes [130].
In Arabidopsis, two HD-Zip I TFs (AtHB7 and AtHB12) have been identified to specifically participate in Al resistance through a reversed genetic approach. Interestingly, AtHB7 and AtHB12 promote root growth through positive regulation of the cell number and cell length under normal conditions, while playing opposite roles by regulating the capacity of the CW to bind Al3+ under Al stress [124]. Recently, HvHOX9, a novel homeobox-leucine zipper transcription factor, was identified to play a critical role in Al tolerance in barley by decreasing root CW Al binding, increasing apoplastic pH in the root, and silencing of HvHOX9 which increased Al accumulation in root CW and decreased H+ influx after Al exposure [136]. Li et al. showed that WRKY47 was involved in altering Al distribution between the apoplast and symplast by regulating the genes responsible for CW modification, thereby improving Al tolerance [125]. Lou et al. reported that a NAC-type TF, VuNAR1, is involved in Al resistance in rice beans, and overexpression of VuNAR1 induced higher WAK1 expression and low pectin content via directly binding to the WAK1 promoter and regulating CW pectin metabolism [138]. Li et al. demonstrated that OsWRKY22 contributes to Al tolerance by functioning together with OsART1 in the positive regulation of OsFRDL4 expression and citrate secretion [126]. A MADS-box transcription factor, GsMAS1, presents a constitutive expression pattern induced under Al stress. The overexpression of GsMAS1 enhanced the tolerance to Al stress in Arabidopsis with larger values of the relative root length and higher proline accumulation as compared with those of wild type (WT) under Al stress through Al stress-related pathways [135]. These findings emphasize the need to study transcription factors involved in Al tolerance, which could help to understand the entire molecular network of Al tolerance in plants, elucidate the mechanism of plant Al tolerance, and lay a theoretical foundation for the cultivation of Al-tolerant varieties using modern molecular techniques.
This entry is adapted from the peer-reviewed paper 10.3390/su13041782
References
- Zheng, S.J. Crop production on acidic soils: Overcoming aluminium toxicity and phosphorus deficiency. Ann. Bot. 2010, 106, 183–184.
- Wei, Y.; Jiang, C.; Han, R.; Xie, Y.; Liu, L.; Yu, Y. Plasma membrane proteomic analysis by TMT-PRM provides insight into mechanisms of aluminum resistance in tamba black soybean roots tips. PeerJ 2020, 8, e9312.
- Kochian, L.V.; Piñeros, M.A.; Liu, J.; Magalhaes, J.V. Plant Adaptation to Acid Soils: The Molecular Basis for Crop Aluminum Resistance. Annu. Rev. Plant Biol. 2015, 66, 571–598.
- Eekhout, T.; Larsen, P.; De Veylder, L. Modification of DNA Checkpoints to Confer Aluminum Tolerance. Trends Plant Sci. 2017, 22, 102–105.
- Yang, L.T.; Qi, Y.P.; Jiang, H.X.; Chen, L.S. Roles of organic acid anion secretion in aluminium tolerance of higher plants. BioMed Res. Int. 2013, 2013, 9–15.
- Jiang, C.; Liu, L.; Li, X.; Han, R.; Wei, Y.; Yu, Y. Insights into aluminum-tolerance pathways in Stylosanthes as revealed by RNA-Seq analysis. Sci. Rep. 2018, 8, 1–9.
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing global ruminant agricultural challenges through understanding the rumen microbiome: Past, present, and future. Front. Microbiol. 2018, 9, 1–33.
- Snowden, K.C.; Cardner, R.C. Five genes induced by aluminum in wheat (Triticum aestivum L.) roots. Plant Physiol. 1993, 103, 855–861.
- Regisfer, P.G.; Richards, K.D.; Snowden, K.C.; Cardner, R.C. wali6 and wali7: Genes induced by aluminum in wheat (Triticum aestivum L.) roots. Plant Physiol. 1994, 105, 1455–1456.
- Muhammad, N.; Zvobgo, G.; Zhang, G.P. A review: The beneficial effects and possible mechanisms of aluminum on plant growth in acidic soil. J. Integr. Agric. 2019, 18, 1518–1528.
- Wang, L.; Fan, X.W.; Pan, J.L.; Huang, Z.B.; Li, Y.Z. Physiological characterization of maize tolerance to low dose of aluminum, highlighted by promoted leaf growth. Planta 2015, 242, 1391–1403.
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266.
- Eticha, D.; Stass, A.; Horst, W.J. Cell-wall pectin and its degree of methylation in the maize root-apex: Significance for genotypic differences in aluminium resistance. Plant Cell Environ. 2005, 28, 1410–1420.
- Singh, S.; Tripathi, D.K.; Singh, S.; Sharma, S.; Dubey, N.K.; Chauhan, D.K.; Vaculík, M. Toxicity of aluminium on various levels of plant cells and organism: A review. Environ. Exp. Bot. 2017, 137, 177–193.
- Wang, Y.; Yu, W.; Cao, Y.; Cai, Y.; Lyi, S.M.; Wu, W.; Kang, Y.; Liang, C.; Liu, J. An exclusion mechanism is epistatic to an internal detoxification mechanism in aluminum resistance in Arabidopsis. BMC Plant Biol. 2020, 20, 1–12.
- Riaz, M.; Yan, L.; Wu, X.; Hussain, S.; Aziz, O.; Jiang, C. Mechanisms of organic acids and boron induced tolerance of aluminum toxicity: A review. Ecotoxicol. Environ. Saf. 2018, 165, 25–35.
- Ma, J.F.; Furukawa, J. Recent progress in the research of external Al detoxification in higher plants: A minireview. J. Inorg. Biochem. 2003, 97, 46–51.
- Lyu, B.; Guo, X.; Gao, D.; Kou, M.; Yu, Y.; Ma, J.; Chen, S.; Wang, H.; Zhang, Y.; Bao, X. Auxin metabolic network regulates the plant response to metalloids stress. J. Hazard. Mater. 2020, 405, 124250.
- Rahman, M.A.; Lee, S.H.; Ji, H.C.; Kabir, A.H.; Jones, C.S.; Lee, K.W. Importance of mineral nutrition for mitigating aluminum toxicity in plants on acidic soils: Current status and opportunities. Int. J. Mol. Sci. 2018, 19, 3073.
- Zhou, J.; Li, Z.; Zhou, T.; Xin, Z.; Wu, L.; Luo, Y.; Christie, P. Aluminum toxicity decreases the phytoextraction capability by cadmium/zinc hyperaccumulator Sedum plumbizincicola in acid soils. Sci. Total Environ. 2020, 711, 134591.
- He, H.; Li, Y.; He, L.F. Aluminum toxicity and tolerance in Solanaceae plants. S. Afr. J. Bot. 2019, 123, 23–29.
- Jaiswal, S.K.; Naamala, J.; Dakora, F.D. Nature and mechanisms of aluminium toxicity, tolerance and amelioration in symbiotic legumes and rhizobia. Biol. Fertil. Soils 2018, 54, 309–318.
- Kopittke, P.M. Role of phytohormones in aluminium rhizotoxicity. Plant Cell Environ. 2016, 39, 2319–2328.
- Sun, L.; Zhang, M.; Liu, X.; Mao, Q.; Shi, C.; Kochian, L.V.; Liao, H. Aluminium is essential for root growth and development of tea plants (Camellia sinensis). J. Integr. Plant Biol. 2020, 62, 984–997.
- Liu, Y.; Tao, J.; Cao, J.; Zeng, Y.; Li, X.; Ma, J.; Huang, Z.; Jiang, M.; Sun, L. The beneficial effects of aluminum on the plant growth in Camellia japonica. J. Soil Sci. Plant Nutr. 2020, 20, 1799–1809.
- Sade, H.; Meriga, B.; Surapu, V.; Gadi, J.; Sunita, M.S.L.; Suravajhala, P.; Kavi Kishor, P.B. Toxicity and tolerance of aluminum in plants: Tailoring plants to suit to acid soils. BioMetals 2016, 29, 187–210.
- Yang, J.l.; Fan, W.; Zheng, S.j. jian Mechanisms and regulation of aluminum-induced secretion of organic acid anions from plant roots. J. Zhejiang Univ. Sci. B 2019, 20, 513–527.
- Qiu, W.; Wang, N.; Dai, J.; Wang, T.; Kochian, L.V.; Liu, J.; Zuo, Y. AhFRDL1-mediated citrate secretion contributes to adaptation to iron deficiency and aluminum stress in peanuts. J. Exp. Bot. 2019, 70, 2873–2886.
- Wu, Y.; Yang, Z.; How, J.; Xu, H.; Chen, L.; Li, K. Overexpression of a peroxidase gene (AtPrx64) of Arabidopsis thaliana in tobacco improves plant’s tolerance to aluminum stress. Plant Mol. Biol. 2017, 95, 157–168.
- Ezaki, B.; Higashi, A.; Nanba, N.; Nishiuchi, T. An S-adenosyl methionine synthetase (SAMS) gene from Andropogon virginicus L. confers aluminum stress tolerance and facilitates epigenetic gene regulation in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 1–13.
- Zhang, L.; Wu, X.X.; Wang, J.; Qi, C.; Wang, X.; Wang, G.; Li, M.; Li, X.; Guo, Y.D. BoALMT1, an al-induced malate transporter in cabbage, enhances aluminum tolerance in Arabidopsis thaliana. Front. Plant Sci. 2018, 8, 1–9.
- Ribeiro, A.P.; de Souza, W.R.; Martins, P.K.; Vinecky, F.; Duarte, K.E.; Basso, M.F.; da Cunha, B.A.D.B.; Campanha, R.B.; de Oliveira, P.A.; Centeno, D.C.; et al. Overexpression of BdMATE gene improves aluminum tolerance in Setaria viridis. Front. Plant Sci. 2017, 8, 1–12.
- Zhou, Y.; Wang, Z.; Gong, L.; Chen, A.; Liu, N.; Li, S.; Sun, H.; Yang, Z.; You, J. Functional characterization of three MATE genes in relation to aluminum-induced citrate efflux from soybean root. Plant Soil 2019, 443, 121–138.
- Ma, Q.; Yi, R.; Li, L.; Liang, Z.; Zeng, T.; Zhang, Y.; Huang, H.; Zhang, X.; Yin, X.; Cai, Z.; et al. GsMATE encoding a multidrug and toxic compound extrusion transporter enhances aluminum tolerance in Arabidopsis thaliana. BMC Plant Biol. 2018, 18, 1–10.
- Li, N.; Meng, H.; Xing, H.; Liang, L.; Zhao, X.; Luo, K. Genome-wide analysis of MATE transporters and molecular characterization of aluminum resistance in Populus. J. Exp. Bot. 2017, 68, 5669–5683.
- Pooniya, V.; Palta, J.A.; Chen, Y.; Delhaize, E.; Siddique, K.H.M. Impact of the TaMATE1B gene on above and below-ground growth of durum wheat grown on an acid and Al3+-toxic soil. Plant Soil 2020, 447, 73–84.
- Xu, J.M.; Lou, H.Q.; Jin, J.F.; Chen, W.W.; Wan, J.X.; Fan, W.; Yang, J.L. A half-type ABC transporter FeSTAR1 regulates Al resistance possibly via UDP-glucose-based hemicellulose metabolism and Al binding. Plant Soil 2018, 432, 303–314.
- Chen, L.; Cai, Y.; Liu, X.; Guo, C.; Yao, W.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. GmGRP-like gene confers Al tolerance in Arabidopsis. Sci. Rep. 2018, 8, 1–12.
- Cai, Z.; Xian, P.; Lin, R.; Cheng, Y.; Lian, T.; Ma, Q.; Nian, H. Characterization of the Soybean GmIREG Family Genes and the Function of GmIREG3 in Conferring Tolerance to Aluminum Stress. Int. J. Mol. Sci. 2020, 21, 497.
- Zhou, Y.; Yang, Z.; Xu, Y.; Sun, H.; Sun, Z.; Lin, B.; Sun, W.; You, J. Soybean NADP-malic enzyme functions in malate and citrate metabolism and contributes to their efflux under Al stress. Front. Plant Sci. 2018, 8, 1–11.
- Li, W.; Du, J.; Feng, H.; Wu, Q.; Xu, G.; Shabala, S.; Yu, L. Function of NHX-type transporters in improving rice tolerance to aluminum stress and soil acidity. Planta 2020, 251, 1–13.
- Liu, W.; Feng, X.; Cao, F.; Wu, D.; Zhang, G.; Vincze, E.; Wang, Y.; Chen, Z.H.; Wu, F. An ATP binding cassette transporter HvABCB25 confers aluminum detoxification in wild barley. J. Hazard. Mater. 2020, 401, 123371.
- Sun, G.; Zhu, H.; Wen, S.; Liu, L.; Gou, L.; Guo, Z. Citrate synthesis and exudation confer Al resistance. Plant Soil 2020, 449, 319–329.
- Li, J.; Su, L.; Lv, A.; Li, Y.; Zhou, P.; An, Y. MsPG1 alleviated aluminum-induced inhibition of root growth by decreasing aluminum accumulation and increasing porosity and extensibility of cell walls in alfalfa (Medicago sativa). Environ. Exp. Bot. 2020, 175, 104045.
- Kariya, K.; Sameeullah, M.; Sasaki, T.; Yamamoto, Y. Overexpression of the sucrose transporter gene NtSUT1 alleviates aluminum-induced inhibition of root elongation in tobacco (Nicotiana tabacum L.). Soil Sci. Plant Nutr. 2017, 63, 45–54.
- Wang, M.; Qiao, J.Y.; Yu, C.L.; Chen, H.; Sun, C.D.; Huang, L.Z.; Li, C.Y.; Geisler, M.; Qian, Q.; Jiang, D.A.; et al. The auxin influx carrier, OsAUX3, regulates rice root development and responses to aluminium stress. Plant Cell Environ. 2019, 42, 1125–1138.
- Lou, H.Q.; Fan, W.; Xu, J.M.; Gong, Y.L.; Jin, J.F.; Chen, W.W.; Liu, L.Y.; Hai, M.R.; Yang, J.L.; Zheng, S.J. An oxalyl-CoA synthetase is involved in oxalate degradation and aluminum tolerance. Plant Physiol. 2016, 172, 1679–1690.
- Lou, H.Q.; Gong, Y.L.; Fan, W.; Xu, J.M.; Liu, Y.; Cao, M.J.; Wang, M.H.; Yang, J.L.; Zheng, S.J. A formate dehydrogenase confers tolerance to aluminum and low pH. Plant Physiol. 2016, 171, 294–305.
- Luo, H.; He, C.; Han, L. Heterologous expression of ZjOMT from Zoysia japonica in Escherichia coli confers aluminum resistance through melatonin production. PLoS ONE 2018, 13, e0196952.
- Du, H.; Huang, Y.; Qu, M.; Li, Y.; Hu, X.; Yang, W.; Li, H.; He, W.; Ding, J.; Liu, C.; et al. A maize ZmAT6 gene confers aluminum tolerance via reactive oxygen species scavenging. Front. Plant Sci. 2020, 11, 1–12.
- Zhang, M.; Lu, X.; Li, C.; Zhang, B.; Zhang, C.; Zhang, X.S.; Ding, Z. Auxin efflux carrier ZmPGP1 mediates root growth inhibition under aluminum stress. Plant Physiol. 2018, 177, 819–832.
- Zhang, X.; Long, Y.; Huang, J.; Xia, J. Molecular mechanisms for coping with al toxicity in plants. Int. J. Mol. Sci. 2019, 20, 1551.
- Pin, M.A.; Magalhaes, J.V.; Alves, V.M.C.; Kochian, L.V. The physiology and biophysics of an aluminum tolerance mechanism based on root citrate exudation in maize. Plant Physiol. 2002, 129, 1194–1206.
- Ligaba, A.; Dreyer, I.; Margaryan, A.; Schneider, D.J.; Kochian, L.; Piñeros, M. Functional, structural and phylogenetic analysis of domains underlying the Al sensitivity of the aluminum-activated malate/anion transporter, TaALMT1. Plant J. 2013, 76, 766–780.
- Hoekenga, O.A.; Maron, L.G.; Piñeros, M.A.; Cançado, G.M.A.; Shaff, J.; Kobayashi, Y.; Ryan, P.R.; Dong, B.; Delhaize, E.; Sasaki, T.; et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 9738–9743.
- Kobayashi, Y.; Hoekenga, O.A.; Itoh, H.; Nakashima, M.; Saito, S.; Shaff, J.E.; Maron, L.G.; Piñeros, M.A.; Kochian, L.V.; Koyama, H. Characterization of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol. 2007, 145, 843–852.
- Gruber, B.D.; Ryan, P.R.; Richardson, A.E.; Tyerman, S.D.; Ramesh, S.; Hebb, D.M.; Howitt, S.M.; Delhaize, E. HvALMT1 from barley is involved in the transport of organic anions. J. Exp. Bot. 2010, 61, 1455–1467.
- Ligaba, A.; Katsuhara, M.; Ryan, P.R.; Shibasaka, M.; Matsumoto, H. The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol. 2006, 142, 1294–1303.
- Ligaba, A.; Katsuhara, M.; Sakamoto, W.; Matsumoto, H. The BnALMT1 protein that is an aluminum-activated malate transporter is localized in the plasma membrane. Plant Signal. Behav. 2007, 2, 255–257.
- Ligaba, A.; Maron, L.; Shaff, J.; Kochian, L.; Piñeros, M. Maize ZmALMT2 is a root anion transporter that mediates constitutive root malate efflux. Plant Cell Environ. 2012, 35, 1185–1200.
- Chen, Z.C.; Yokosho, K.; Kashino, M.; Zhao, F.J.; Yamaji, N.; Ma, J.F. Adaptation to acidic soil is achieved by increased numbers of cis-acting elements regulating ALMT1 expression in Holcus lanatus. Plant J. 2013, 76, 10–23.
- Park, W.; Kim, H.S.; Park, T.W.; Lee, Y.H.; Ahn, S.J. Functional characterization of plasma membrane-localized organic acid transporter (CsALMT1) involved in aluminum tolerance in Camelina sativa L. Plant Biotechnol. Rep. 2017, 11, 181–192.
- Ma, X.; An, F.; Wang, L.; Guo, D.; Xie, G.; Liu, Z. Genome-wide identification of aluminum-activated malate transporter (ALMT) gene family in rubber trees (Hevea brasiliensis) highlights their involvement in aluminum detoxification. Forests 2020, 11, 142.
- Upadhyay, N.; Kar, D.; Deepak Mahajan, B.; Nanda, S.; Rahiman, R.; Panchakshari, N.; Bhagavatula, L.; Datta, S. The multitasking abilities of MATE transporters in plants. J. Exp. Bot. 2019, 70, 4643–4656.
- Liu, J.; Magalhaes, J.V.; Shaff, J.; Kochian, L.V. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 2009, 57, 389–399.
- Wu, X.; Li, R.; Shi, J.; Wang, J.; Sun, Q.; Zhang, H.; Xing, Y.; Qi, Y.; Zhang, N.; Guo, Y.D. Brassica oleracea MATE encodes a citrate transporter and enhances aluminum tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 1426–1436.
- Sawaki, Y.; Kihara-Doi, T.; Kobayashi, Y.; Nishikubo, N.; Kawazu, T.; Kobayashi, Y.; Koyama, H.; Sato, S. Characterization of Al-responsive citrate excretion and citrate-transporting MATEs in Eucalyptus camaldulensis. Planta 2013, 237, 979–989.
- Lei, G.J.; Yokosho, K.; Yamaji, N.; Ma, J.F. Two MATE transporters with different subcellular localization are involved in Al tolerance in buckwheat. Plant Cell Physiol. 2017, 58, 2179–2189.
- Liu, J.; Li, Y.; Wang, W.; Gai, J.; Li, Y. Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genom. 2016, 17, 1–15.
- Wang, P.; Yu, W.; Zhang, J.; Rengel, Z.; Xu, J.; Han, Q.; Chen, L.; Li, K.; Yu, Y.; Chen, Q. Auxin enhances aluminium-induced citrate exudation through upregulation of GmMATE and activation of the plasma membrane H+-ATPase in soybean roots. Ann. Bot. 2016, 118, 933–940.
- Iguchi, A.; Sanmiya, K.; Watanabe, K. Identification of genes encoding ALMT and MATE transporters as candidate aluminum tolerance genes from a typical acid soil plant, Psychotria rubra (Rubiaceae). PeerJ 2019, 7, e7739.
- Melo, J.O.; Lana, U.G.P.; Piñeros, M.A.; Alves, V.M.C.; Guimarães, C.T.; Liu, J.; Zheng, Y.; Zhong, S.; Fei, Z.; Maron, L.G.; et al. Incomplete transfer of accessory loci influencing SbMATE expression underlies genetic background effects for aluminum tolerance in sorghum. Plant J. 2013, 73, 276–288.
- Liu, M.Y.; Chen, W.W.; Xu, J.M.; Fan, W.; Yang, J.L.; Zheng, S.J. The role of VuMATE1 expression in aluminium-inducible citrate secretion in rice bean (Vigna umbellata) roots. J. Exp. Bot. 2013, 64, 1795–1804.
- Fan, W.; Lou, H.Q.; Gong, Y.L.; Liu, M.Y.; Cao, M.J.; Liu, Y.; Yang, J.L.; Zheng, S.J. Characterization of an inducible C2H2-type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and aluminum tolerance mechanisms. New Phytol. 2015, 208, 456–468.
- Yang, X.Y.; Yang, J.L.; Zhou, Y.; Piñeros, M.A.; Kochian, L.V.; Li, G.X.; Zheng, S.J. A de novo synthesis citrate transporter, Vigna umbellata multidrug and toxic compound extrusion, implicates in Al-activated citrate efflux in rice bean (Vigna umbellata) root apex. Plant Cell Environ. 2011, 34, 2138–2148.
- Liu, M.Y.; Lou, H.Q.; Chen, W.W.; Piñeros, M.A.; Xu, J.M.; Fan, W.; Kochian, L.V.; Zheng, S.J.; Yang, J.L. Two citrate transporters coordinately regulate citrate secretion from rice bean root tip under aluminum stress. Plant Cell Environ. 2018, 41, 809–822.
- Maron, L.G.; Piñeros, M.A.; Guimarães, C.T.; Magalhaes, J.V.; Pleiman, J.K.; Mao, C.; Shaff, J.; Belicuas, S.N.J.; Kochian, L.V. Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J. 2010, 61, 728–740.
- Zhu, H.; Wu, J.; Jiang, Y.; Jin, J.; Zhou, W.; Wang, Y.; Han, G.; Zhao, Y.; Cheng, B. Genomewide analysis of MATE-type gene family in maize reveals microsynteny and their expression patterns under aluminum treatment. J. Genet. 2016, 95, 691–704.
- Semenova, E.V.; Kosareva, I.A.; Belimov, A.A. Aluminum exclusion from root zone and maintenance of nutrient uptake are principal mechanisms of Al tolerance in Pisum sativum L. Physiol. Mol. Biol. Plants 2017, 23, 851–863.
- Klug, B.; Horst, W.J. Oxalate exudation into the root-tip water free space confers protection from aluminum toxicity and allows aluminum accumulation in the symplast in buckwheat (Fagopyrum esculentum). New Phytol. 2010, 187, 380–391.
- Zhang, J.; Wei, J.; Li, D.; Kong, X.; Rengel, Z.; Chen, L.; Yang, Y.; Cui, X.; Chen, Q. The role of the plasma membrane H+-ATPase in plant responses to aluminum toxicity. Front. Plant Sci. 2017, 8, 1–9.
- Yang, Y.; Wang, Q.L.; Geng, M.J.; Guo, Z.H.; Zhao, Z. Rhizosphere ph difference regulated by plasma membrane H+-ATPase is related to differential Al-tolerance of two wheat cultivars. Plant Soil Environ. 2011, 57, 201–206.
- Zhang, F.; Yan, X.; Han, X.; Tang, R.; Chu, M.; Yang, Y.; Yang, Y.H.; Zhao, F.; Fu, A.; Luan, S.; et al. A defective vacuolar proton pump enhances aluminum tolerance by reducing vacuole sequestration of organic acids. Plant Physiol. 2019, 181, 743–761.
- Kidd, P.S.; Llugany, M.; Poschenrieder, C.; Gunsé, B.; Barceló, J. The role of root exudates in aluminium resistance and silicon-induced amelioration of aluminium toxicity in three varieties of maize (Zea mays L.). J. Exp. Bot. 2001, 52, 1339–1352.
- Ma, Z.; Lin, S. Transcriptomic revelation of phenolic compounds involved in aluminum toxicity responses in roots of cunninghamia lanceolata (lamb.) hook. Genes 2019, 10, 835.
- Chen, Y.; Huang, L.; Liang, X.; Dai, P.; Zhang, Y.; Li, B.; Lin, X.; Sun, C. Enhancement of polyphenolic metabolism as an adaptive response of lettuce (Lactuca sativa) roots to aluminum stress. Environ. Pollut. 2020, 261, 114230.
- Fu, Z.; Jiang, X.; Li, W.W.; Shi, Y.; Lai, S.; Zhuang, J.; Yao, S.; Liu, Y.; Hu, J.; Gao, L.; et al. Proanthocyanidin-aluminum complexes improve aluminum resistance and detoxification of Camellia sinensis. J. Agric. Food Chem. 2020, 68, 7861–7869.
- Yang, J.L.; Li, Y.Y.; Zhang, Y.J.; Zhang, S.S.; Wu, Y.R.; Wu, P.; Zheng, S.J. Cell wall polysaccharides are specifically involved in the exclusion of aluminum from the rice root apex. Plant Physiol. 2008, 146, 602–611.
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 2009, 21, 655–667.
- Che, J.; Yamaji, N.; Yokosho, K.; Shen, R.F.; Ma, J.F. Two genes encoding a bacterial-type ATP-binding cassette transporter are implicated in aluminum tolerance in buckwheat. Plant Cell Physiol. 2018, 59, 2502–2511.
- Fan, W.; Xu, J.M.; Wu, P.; Yang, Z.X.; Lou, H.Q.; Chen, W.W.; Jin, J.F.; Zheng, S.J.; Yang, J.L. Alleviation by abscisic acid of Al toxicity in rice bean is not associated with citrate efflux but depends on ABI5-mediated signal transduction pathways. J. Integr. Plant Biol. 2019, 61, 140–154.
- Yang, T.-Y.; Huang, W.-T.; Zhang, J.; Yang, L.-T.; Huang, Z.-R.; Wu, B.-S.; Lai, N.-W.; Chen, L.-S. Raised pH conferred the ability to maintain a balance between production and detoxification of reactive oxygen species and methylglyoxal in aluminum-toxic Citrus sinensis leaves and roots. Environ. Pollut. 2020, 268, 115676.
- Yang, X.Y.; Zeng, Z.H.; Yan, J.Y.; Fan, W.; Bian, H.W.; Zhu, M.Y.; Yang, J.L.; Zheng, S.J. Association of specific pectin methylesterases with Al-induced root elongation inhibition in rice. Physiol. Plant. 2013, 148, 502–511.
- Baylis, A.D.; Gragopoulou, C.; Davidson, K.J.; Birchall, J.D. Effects of silicon on the toxicity of aluminium to soybean. Commun. Soil Sci. Plant Anal. 1994, 25, 537–546.
- Kochian, L.V. Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 237–260.
- Yang, J.L.; Zhu, X.F.; Peng, Y.X.; Zheng, C.; Li, G.X.; Liu, Y.; Shi, Y.Z.; Zheng, S.J. Cell wall hemicellulose contributes significantly to aluminum adsorption and root growth in Arabidopsis. Plant Physiol. 2011, 155, 1885–1892.
- Yang, T.Y.; Cai, L.Y.; Qi, Y.P.; Yang, L.T.; Lai, N.W.; Chen, L.S. Increasing nutrient solution ph alleviated aluminum-induced inhibition of growth and impairment of photosynthetic electron transport chain in Citrus sinensis seedlings. BioMed Res. Int. 2019, 2019, 9058715.
- Yang, T.Y.; Qi, Y.P.; Huang, H.Y.; Wu, F.L.; Huang, W.T.; Deng, C.L.; Yang, L.T.; Chen, L.S. Interactive effects of pH and aluminum on the secretion of organic acid anions by roots and related metabolic factors in Citrus sinensis roots and leaves. Environ. Pollut. 2020, 262, 114303.
- Kong, X.; Peng, Z.; Li, D.; Ma, W.; An, R.; Khan, D.; Wang, X.; Liu, Y.; Yang, E.; He, Y.; et al. Magnesium decreases aluminum accumulation and plays a role in protecting maize from aluminum-induced oxidative stress. Plant Soil 2020, 457, 71–81.
- Zhang, Z.; Liu, D.; Meng, H.; Li, S.; Wang, S.; Xiao, Z.; Sun, J.; Chang, L.; Luo, K.; Li, N. Magnesium alleviates aluminum toxicity by promoting polar auxin transport and distribution and root alkalization in the root apex in populus. Plant Soil 2020, 448, 565–585.
- Li, J.Y.; Liu, J.; Dong, D.; Jia, X.; McCouch, S.R.; Kochian, L.V. Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. Proc. Natl. Acad. Sci. USA 2014, 111, 6503–6508.
- Xia, J.; Yamaji, N.; Kasai, T.; Ma, J.F. Plasma membrane-localized transporter for aluminum in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 18381–18385.
- Nhan, P. Amelioration of aluminum toxicity on OM4900 rice seedlings by sodium silicate. Afr. J. Plant Sci. 2013, 7, 208–212.
- Carr, H.P.; Lombi, E.; Küpper, H.; McGrath, S.P.; Wong, M.H. Accumulation and distribution of aluminium and other elements in tea (Camellia sinensis) leaves. Agronomie 2003, 23, 705–710.
- Negishi, T.; Oshima, K.; Hattori, M.; Kanai, M.; Mano, S.; Nishimura, M.; Yoshida, K. Tonoplast-and plasma membrane-localized aquaporin-family transporters in Blue Hydrangea Sepals of Aluminum Hyperaccumulating Plant. PLoS ONE 2012, 7, e43189.
- Lu, M.; Yang, G.; Li, P.; Wang, Z.; Fu, S.; Zhang, X.; Chen, X.; Shi, M.; Ming, Z.; Xia, J. Bioinformatic and functional analysis of a key determinant underlying the substrate selectivity of the al transporter, Nrat1. Front. Plant Sci. 2018, 9, 1–9.
- Larsen, P.B.; Geisler, M.J.B.; Jones, C.A.; Williams, K.M.; Cancel, J.D. ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J. 2005, 41, 353–363.
- Lei, G.J.; Yokosho, K.; Yamaji, N.; Fujii-Kashino, M.; Ma, J.F. Functional characterization of two half-size ABC transporter genes in aluminium-accumulating buckwheat. New Phytol. 2017, 215, 1080–1089.
- Ezaki, B.; Takahashi, K.; Utsumi, K.; Higashi, A. A half-type AvABCG1 transporter derived from Andropogon virginicus L. confers aluminum tolerance. Environ. Exp. Bot. 2015, 118, 21–31.
- Wang, Y.; Li, R.; Li, D.; Jia, X.; Zhou, D.; Li, J.; Lyi, S.M.; Hou, S.; Huang, Y.; Kochian, L.V.; et al. NIP1;2 is a plasma membrane-localized transporter mediating aluminum uptake, translocation, and tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 5047–5052.
- Hazak, O.; Mamon, E.; Lavy, M.; Sternberg, H.; Behera, S.; Schmitz-Thom, I.; Bloch, D.; Dementiev, O.; Gutman, I.; Danziger, T.; et al. A novel Ca2+-binding protein that can rapidly transduce auxin responses during root growth. PLoS Boil. 2019, 17, e3000085.
- Kar, D.; Pradhan, A.A.; Datta, S. The role of solute transporters in aluminium toxicity and tolerance. Physiol. Plant. 2020.
- Lan, T.; You, J.; Kong, L.; Yu, M.; Liu, M.; Yang, Z. The interaction of salicylic acid and Ca2+ alleviates aluminum toxicity in soybean (Glycine max L.). Plant Physiol. Biochem. 2016, 98, 146–154.
- Su, L.; Lv, A.; Wen, W.; Zhou, P.; An, Y. Auxin is involved in magnesium-mediated photoprotection in photosystems of alfalfa seedlings under aluminum stress. Front. Plant Sci. 2020, 11, 1–17.
- Chen, Z.C.; Yamaji, N.; Motoyama, R.; Nagamura, Y.; Ma, J.F. Up-regulation of a magnesium transporter gene OsMGT1 is required for conferring aluminum tolerance in rice. Plant Physiol. 2012, 159, 1624–1633.
- Li, D.; Ma, W.; Wei, J.; Mao, Y.; Peng, Z.; Zhang, J.; Kong, X.; Han, Q.; Fan, W.; Yang, Y.; et al. Magnesium promotes root growth and increases aluminum tolerance via modulation of nitric oxide production in Arabidopsis. Plant Soil 2019, 1–13.
- Vera-Villalobos, H.; Lunario-Delgado, L.; Pérez-Retamal, D.; Román, D.; Leiva, J.C.; Zamorano, P.; Mercado-Seguel, A.; Gálvez, A.S.; Benito, C.; Wulff-Zottele, C. Sulfate nutrition improves short-term Al3+-stress tolerance in roots of Lolium perenne L. Plant Physiol. Biochem. 2020, 148, 103–113.
- Yamamoto, Y.; Kobayashi, Y.; Devi, S.R.; Rikiishi, S.; Matsumoto, H. Oxidative stress triggered by aluminum in plant roots. Plant Soil 2003, 255, 239–243.
- Sun, C.; Liu, L.; Zhou, W.; Lu, L.; Jin, C.; Lin, X. Aluminum induces distinct changes in the metabolism of reactive oxygen and nitrogen species in the roots of two wheat genotypes with different aluminum resistance. J. Agric. Food Chem. 2017, 65, 9419–9427.
- Basu, U.; Good, A.G.; Taylor, G.J. Transgenic Brassica napus plants overexpressing aluminium-induced mitochondrial manganese superoxide dismutase cDNA are resistant to aluminium. Plant Cell Environ. 2001, 24, 1269–1278.
- Ezaki, B.; Gardner, R.C.; Ezaki, Y.; Matsumoto, H. Expression of aluminum-induced genes in transgenic Arabidopsis plants can ameliorate aluminum stress and/or oxidative stress. Plant Physiol. 2000, 122, 657–665.
- Guo, Y.; Qiao, D.; Yang, C.; Chen, J.; Li, Y.; Liang, S.; Lin, K.; Chen, Z. Genome-wide identification and expression analysis of SABATH methyltransferases in tea plant (Camellia sinensis): Insights into their roles in plant defense responses. Plant Signal. Behav. 2020, 15, 1804684.
- Jin, J.F.; Wang, Z.Q.; He, Q.Y.; Wang, J.Y.; Li, P.F.; Xu, J.M.; Zheng, S.J.; Fan, W.; Yang, J.L. Genome-wide identification and expression analysis of the NAC transcription factor family in tomato (Solanum lycopersicum) during aluminum stress. BMC Genom. 2020, 21, 1–14.
- Liu, Y.; Xu, J.; Guo, S.; Yuan, X.; Zhao, S.; Tian, H.; Dai, S.; Kong, X.; Ding, Z. AtHB7/12 regulate root growth in response to aluminum stress. Int. J. Mol. Sci. 2020, 21, 4080.
- Li, C.X.; Yan, J.Y.; Ren, J.Y.; Sun, L.; Xu, C.; Li, G.X.; Ding, Z.J.; Zheng, S.J. A WRKY transcription factor confers aluminum tolerance via regulation of cell wall modifying genes. J. Integr. Plant Biol. 2019, 62, 1176–1192.
- Li, G.Z.; Wang, Z.Q.; Yokosho, K.; Ding, B.; Fan, W.; Gong, Q.Q.; Li, G.X.; Wu, Y.R.; Yang, J.L.; Ma, J.F.; et al. Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4 expression and enhancement of citrate secretion in rice (Oryza sativa). New Phytol. 2018, 219, 149–162.
- Arun, A.; Yuriko, D.; Sanjib, K.; Panda, K. Characterization of CcSTOP1; a C2H2-type transcription factor regulates Al tolerance gene in pigeonpea. Planta 2017, 247, 201–214.
- Kundu, A.; Das, S.; Basu, S.; Kobayashi, Y.; Kobayashi, Y.; Koyama, H.; Ganesan, M. GhSTOP1, a C2H2 type zinc finger transcription factor is essential for aluminum and proton stress tolerance and lateral root initiation in cotton. Plant Biol. 2019, 21, 35–44.
- Zhou, Y.; Yang, Z.M.; Gong, L.; Liu, R.K.; Sun, H.R.; You, J.F. Molecular characterization of GmSTOP1 homologs in soybean under Al and proton stress. Plant Soil 2018, 427, 213–230.
- Liu, Y.T.; Shi, Q.H.; Cao, H.J.; Ma, Q.B.; Nian, H.; Zhang, X.X. Heterologous expression of a Glycine soja C2H2 zinc finger gene improves aluminum tolerance in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 2754.
- Wu, L.; Guoa, Y.; Caia, S.; Kuanga, L.; Shena, Q.; Wu, D.; Zhanga, G. A zinc finger transcription factor HvATF1 regulates aluminum tolerance in barley. J. Exp. Bot. 2020, 71, 6512–6523.
- Ito, H.; Kobayashi, Y.; Yamamoto, Y.Y.; Koyama, H. Characterization of NtSTOP1-regulating genes in tobacco under aluminum stress. Soil Sci. Plant Nutr. 2019, 65, 251–258.
- Che, J.; Tsutsui, T.; Yokosho, K.; Yamaji, N.; Ma, J.F. Functional characterization of an aluminum (Al)-inducible transcription factor, ART2, revealed a different pathway for Al tolerance in rice. New Phytol. 2018, 220, 209–218.
- Huang, S.; Gao, J.; You, J.; Liang, Y.; Guan, K.; Yan, S.; Zhan, M.; Yang, Z. Identification of STOP1-like proteins associated with aluminum tolerance in sweet sorghum (Sorghum bicolor L.). Front. Plant Sci. 2018, 9, 258.
- Zhang, X.; Li, L.; Yang, C.; Cheng, Y.; Han, Z.; Cai, Z.; Nian, H.; Ma, Q. GsMAS1 encoding a MADS-box transcription factor enhances the tolerance to aluminum stress in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 2004.
- Feng, X.; Liu, W.; Dai, H.; Qiu, Y.; Zhang, G.; Chen, Z.-H.; Wu, F. HvHOX9, a novel homeobox leucine zipper transcription factor revealed by root miRNA and RNA sequencing in tibetan wild barley, positively regulates Al tolerance. J. Exp. Bot. 2020, 71, 6057–6073.
- An, J.P.; Wang, X.N.; Yao, J.F.; Ren, Y.R.; You, C.X.; Wang, X.F.; Hao, Y.J. Apple MdMYC2 reduces aluminum stress tolerance by directly regulating MdERF3 gene. Plant Soil 2017, 418, 255–266.
- Lou, H.Q.; Fan, W.; Jin, J.F.; Xu, J.M.; Chen, W.W.; Yang, J.L.; Zheng, S.J. A NAC-type transcription factor confers aluminium resistance by regulating cell wall-associated receptor kinase 1 and cell wall pectin. Plant. Cell Environ. 2020, 43, 463–478.
- Zhang, Y.; Zhang, J.; Guo, J.; Zhou, F.; Singh, S.; Xu, X.; Xie, Q.; Yang, Z.; Huang, C.F. F-box protein RAE1 regulates the stability of the aluminum-resistance transcription factor STOP1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 319–327.
- Guo, J.; Zhang, Y.; Gao, H.; Li, S.; Wang, Z.-Y.; Huang, C.-F. Mutation of HPR1 encoding a component of the THO/TREX complex reduces STOP1 accumulation and aluminium resistance in Arabidopsis thaliana. New Phytol. 2020, 228, 179–193.
- Fang, Q.; Zhang, J.; Zhang, Y.; Fan, N.; van den Burg, H.A.; Huang, C.F. Regulation of aluminum-resistance in Arabidopsis involves the SUMOylation of the zinc finger transcription factor STOP1. Plant Cell 2020, 32, 3921–3938.
- Tsutsui, T.; Yamaji, N.; Feng Ma, J. Identification of a cis-acting element of ART1, a C2H2-type zinc-finger transcription factor for aluminum tolerance in rice. Plant Physiol. 2011, 156, 925–931.
