Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Chemistry, Medicinal
In the current era, an ever-emerging threat of multidrug-resistant (MDR) pathogens pose serious health challenges to mankind. Researchers are uninterruptedly putting their efforts to design and develop alternative, innovative strategies to tackle the antibiotic resistance displayed by varied pathogens. Among several naturally derived and chemically synthesized compounds, quinones have achieved a distinct position to defeat microbial pathogens.
- efflux pumps
- MDR
- ESKAPE pathogens
- naphthoquinones
- plasmid curing
- reactive oxygen species
- topoisomerase
Note: The following contents are extract from your paper. The entry will be online only after author check and submit it.
1. Introduction
Antibiotics represent world-class, assured molecules that have captured a gigantic share in the global market to combat ever-rising and prevalent infections. Over decades, different types of antibiotics have come into medical practice. Penicillin occupied the European and U.S. markets since its discovery in 1928 by Sir Alexander Fleming, followed by its commercial production in the 1940s [1]. Further, the world was gifted with the discovery of another antibiotic, streptomycin, by Albert Schatz, Bugie, and Waksman in 1943. This antibiotic was able to inhibit bacteria, predominantly the organisms responsible for tuberculosis [2]. After the success stories of penicillin and streptomycin, a huge number of antibiotics succeeded commercially, such that the projected rise of the global antibiotic market is up to US $67.25 billion by 2026 [3].
The challenges of resistance acquired by microbial pathogens towards existing antibiotics led to the advent of new antimicrobials. Presently, healthcare sectors are severely affected due to eternally escalating antimicrobial resistance (AMR) shown by pathogenic bacteria, parasites, viruses, and fungi. This serious threat associated with public health needs urgent attention and an immediate action plan from government policymakers, as well as private industries. It is essential to note that the challenges associated with AMR have led to a substantial cost escalation for pharmaceutical and health-care products. Patients suffering from microbial infections are ultimately the victims of long-term illness, and are therefore loaded with an additional monetary burden in the form of expensive tests and drugs [4,5]. There is also an increased morbidity and mortality rate in patients. These multidrug-resistant (MDR) pathogens are therefore referred to as “Super Bugs” [6]. MDR bacterial pathogens also comprise the ESKAPE group [7]. The abbreviation ‘ESKAPE’ has been used to designate a group having Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacterspp. In the year 2008, Rice [8] had coined the terminology ‘the ESKAPE bugs’ to denote the ability of pathogens to escape from the lethal activity of antibiotics and impose severe menace to human health. These pathogens exhibit resistance to antimicrobial drugs like carbapenems, fluoroquinolones, glycopeptides, β-lactams, β-lactam–β-lactamase inhibitor combinations, lipopeptides, macrolides, tetracyclines, and polymyxins etc. [7]. In the year 2017, the World Health Organization (WHO) published a list of priority pathogens—Priority 1 (Critical), Priority 2 (High) and Priority 3 (Medium)—exhibiting resistance to antimicrobial agents [9]. These pathogens can worsen emergency situations and therefore, need urgent surveillance so that new and more effective compounds can be brought through the pipelines. Since 2015, WHO has introduced World Antibiotic Awareness Week (WAAW) to create awareness in the public community, health workforce and among policy-formulating personnel to restrict further emergence of antibiotic resistance and its spread. Since 2020, the Tripartite Executive Committee declared WAAW dates to be 18–24 November [10].
To gain AMR, pathogens acquire plasmids (R-drug resistance) or transposons and also possess multidrug efflux pumps (EPs) to drive out the drug molecules from their system [11]. Besides, other strategies are used, like (1) inactivation, alteration, or degradation of the drug by bacterial enzymes, (2) modification of drug binding sites on the bacterial cell, (3) biofilm formation (restricting the entry of the drug), and (4) reduction in intracellular drug accumulation [12]. Challenges associated with AMR encouraged researchers to explore a variety of naturally existing and chemically synthesized compounds over the decades. Since ancient times, medicinal plants have been evidenced as a great support to tackle dreaded illnesses [13]. Recent advances in the area of phytochemicals and synthesized derivatives have been looked forward to due to their multifunctional therapeutic approaches for dealing with AMR-associated challenges [14,15,16,17]. The unique structural, biological, and functional properties of naphthoquinones (NQs), along with their derivatives, have gained enormous consideration, particularly from a medicinal chemistry perspective [18]. NQs are widely distributed as natural pigments in plants, fungi, and some animals [19]. NQ derivatives bearing hydroxyl, methyl, nitrogen, sulfur, halide, phenylamino-phenylthio, or sulphide possess exceptional biological activity. Derivatives bearing hydroxyl groups are seeking more consideration due to their broad-spectrum pharmacological properties [20]. NQs possess widespread antibacterial, antiparasitic [21,22], antifungal, [23,24], antiviral [25], and antimalarial properties [26]. In the field of cancer biology, NQs are also noticeably identified for their abilities to produce reactive oxygen species (ROS) in cancer cell lines [27,28,29]. NQs are propitious candidates over the other chemotherapeutic drugs used currently. Until today, an ample number of NQ derivatives have been analyzed for their functional potential against various pathogens. This encouraged us to present a comprehensive review of the structural diversity and multifunctional potentiality of NQs in medicinal chemistry. We also shed a light on the current understanding of the mechanistic roles of NQs in combating microbial infections. We also emphasize molecular docking—a powerful approach used in predicting the interactions of NQ molecules in biological systems.
2. Structural Diversity of Naphthoquinones Entities
Structurally, NQs constitute bicyclic structures with two carbonyl groups placed either in positions 1, 4 or 1, 2. The latter case is less frequent (Figure 1A). The chemistry and biological activities of quinones are primarily dependent on the position and chemical nature of the side groups attached (R). Groups like hydrogen, hydroxyl, methyl, nitrogen, sulfur, halide, etc. are attached to the NQ’s ring structure. Generally, the presence of a hydroxyl and/or methyl group in quinone structure is found in nature. These derivatives have been reported for a broad range of applications in pharmacology [19]. Recently, NQ derivatives isolated from plant sources including lawsone, juglone, plumbagin, shikonin and lapachol (Table 1) have fascinated researchers due to their (1) abundance, (2) structural diversity, and (3) broad-spectrum therapeutic potential [20]. It is imperative to state that 1,4-NQs derivatized at the 2nd and 3rd position with different chemical groups are recurrently reported for their biological properties (Table 1). NQs having oxygen entities at 1, 4 positions in the aromatic ring exhibit promising antimicrobial properties. Sparse literature is evident on 1,2-NQs. Realizing the significance of 1,4-NQ derivatives for biological applications, the major focus of this review remains on 1,4-NQ derivatives.
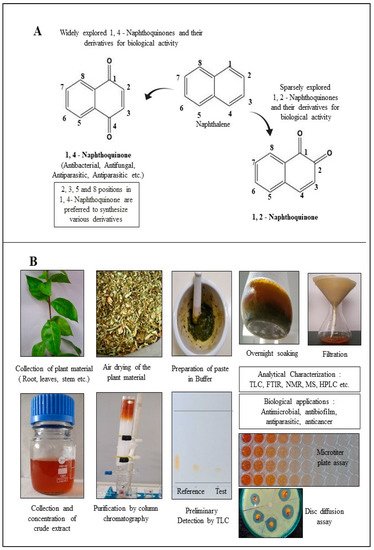
Figure 1. Diversity of naphthoquinone molecules. (A): structures of chemically synthesized naphthoquinones. (B): production, purification, characterization, and biological applications of 1,4-naphthoquinone derivatives from plant material.
Table 1. Chemical structures of plant-originated naphthoquinone derivatives.
| Name and Structure of the Naphthoquinone Derivative | Source of Plant Material | Reference |
|---|---|---|
(A) Juglone: 5-Hydroxy-1,4-naphthalenedione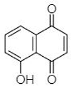 |
Caesalpinia sappan | [30] |
(B) Plumbagin: 5-Hydroxy-2-methyl-naphthalene-1,4-dione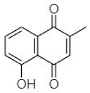 |
Plumbago zeylanica | [31] |
| Plumbago auriculata | [32] | |
| Plumbago indica | [33] | |
(C) Lawsone: 2-Hydroxy-1,4-naphthoquinone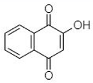 |
Plumbago zeylanica | [34] |
| Lawsonia inermis | [35] | |
(D) Shikonin: 5,8-Dihydroxy-2-[(1R)-1-hydroxy-4-methyl-3-penten-1-yl]-1,4-naphthoquinone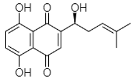 |
Arnebia euchroma | [36] |
| Lithospermum erythrorhizon | [37] | |
(E) Lapachol: 2-Hydroxy-3-(3-methyl-2-butenyl)-1,4-Naphthalenedione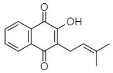 |
Tabebuia ochracea | [38,39] |
3. 1,4-Naphthoquinone Derivatives
The 1,4-NQs are redox-active compounds having resemblance to the structure of naphthalene [40]. Researchers have focused on 1,4-NQ derivatives to explore for antibacterial, antifungal, antiviral, anticancer, and antiparasitic potential. The following sections describe 1,4-NQs, both natural-origin and chemically synthesized, having immense antimicrobial potential against various pathogens, including those that are MDR or in the ESKAPE category.
3.1. 1,4-Naphthoquinone Derivatives of Natural Origin
Naphthoquinones, phenol-rich compounds, are seen abundantly in a variety of plants [30,32], animals [41], and fungi [42]. The functional potency of any antimicrobial agent is uniquely determined from its minimum inhibitory concentration (MIC). The MIC represents the lowest concentration that hampers the visible growth of microbes. Minimum bactericidal concentrations (MBCs) indicate the lowest concentration of a compound that prevents the growth of an organism by killing it. Therefore, MICs and MBCs are imperative for any antimicrobial agent to be considered for pharmacological applications. MICs of plant-derived NQs against pathogenic bacteria are represented in Table 2.
Table 2. Naphthoquinones extracted from plants along with their minimum inhibitory concentration (MIC).
| Type of Naphthoquinone | Active against Bacterial Strains | MIC (µg/mL) | Reference |
|---|---|---|---|
| Shikonin from Arnebia euchroma root extract | Staphylococcus aureus | 128 | [36] |
| Streptococcus agalactiae | 128 | ||
| Escherichia coli | 256 | ||
| Salmonella isolates | 256 | ||
| Pseudomonas aeruginosa | 512 | ||
| Plumbagin from Plumbago zeylanica | Staphylococcus aureus | 0.5 | [43] |
| Escherichia coli | 8 | ||
| Klebsiella pneumoniae | 2 | ||
| Pseudomonas aeruginosa | 8 | ||
| Plumbagin from Plumbago zeylanica | S. aureus (MRSA) | 4–8 | [31] |
| Lawsone from Plumbago zeylanica root extract | Staphylococcus aureus | 400 | [34] |
| Salmonella typhi | 200 | ||
| Bacillus cereus | 200 | ||
| Bacillus subtilis | 200 | ||
| Pseudomonas aeruginosa | 800 | ||
| Escherichia coli | 800 | ||
| Shigella dysenteriae | 400 | ||
| Serratia marcescens | >1600 | ||
| Proteus mirabilis | >1600 | ||
| Klebsiellapneumoniae | 800 | ||
| Enterobacter | 800 | ||
| Acinetobacter baumannii | 800 | ||
| Naphthoquinone pigments from Onosma visianii | Bacillus megaterium | 9.54–54.28 | [44] |
| Montrichardia arborescens | 6.82–54.28 | ||
| Micrococcus luteus | 9.54–76.20 | ||
| Staphylococcus epidermidis | 9.54–54.28 | ||
| Enterococcus faecalis | 6.82–38.10 | ||
| Citrobacter koseri | 6.82–76.20 | ||
| Hafnia alvei | 6.82–54.28 | ||
| Pseudomonas proteolytica | 4.77–76.20 | ||
| Stenotrophomonas maltophilia | 4.77–76.20 | ||
| Yersinia intermedia | 6.82–76.20 | ||
| - | (values depict MIC90) | ||
| Shikonin from Lithospermum erythrorhizon | Staphylococcus aureus (MRSA) | 7.8–31.2 | [37] |
| Staphylococcus aureus (MSSA) | 7.8 | ||
| Plumbago auriculata root extracts | Proteus vulgaris | Strong activity | [32] |
| Klebsiella. pneumoniae | Strong activity | ||
| Escherichia coli | Moderate activity | ||
| Pseudomonas aeruginosa | Less activity | ||
| Naphthoquinone from Alkanna orientalis extracts | Bacillus megaterium | 125 | [45] |
| Bacillus mesentericus | 125 | ||
| Bacillus mycoides | 125 | ||
| Bacillus subtilis | 125 | ||
| Brevibacterium flavum | 250 | ||
| Enterococcus hirae | 250 | ||
| Micrococcus luteus | 250 | ||
| Staphylococcus aureus | 125 | ||
| Staphylococcus citreus | 500 | ||
| Staphylococcus roseus | 125 | ||
| Escherichia. coli | 500 | ||
| Salmonella typhimurium | 750 | ||
| Lawsone from Lawsonia inermis | Bacillus subtilis | Activity at 1000 µg/disc | [35] |
| Staphylococcus aureus | No activity | ||
| Escherichia coli | No activity | ||
| Pseudomonas aeruginosa | No activity | ||
| Plumbago indica from root extracts | Cutibacterium acnes (formerly Propionibacterium acnes) | 12.5 | [33] |
| Staphylococcus aureus | 12.5 | ||
| Staphylococcus epidermidis | 0.78 | ||
| Naphthoquinones from Caesalpinia sappan | Clostridium perfringens | Activity observed | [30] |
| Bacillus bifidum | Activity observed | ||
| Bacillus. breve | No significant activity | ||
| Lactobacillus casei | Minute activity | ||
| Escherichia coli | Minute activity |
This section is devoted to the structural diversity and functional potentiality of NQs of plant origin. Various plants, namely, Plumbago zeylanica, Plumbago indica, Lawsonia inermis, Plumbago auriculata, Caesalpinia sappan, Onosma visianii, and Impatiens glandulifera, and solvents like methanol, chloroform, acetone, ethanol, petroleum ether, diethyl ether, benzene, acetonitrile (ACN), dimethyl sulfoxide (DMSO), dimethylformamide (DMF), ethyl acetate (EA), isopropyl alcohol (IPA), and water etc. have been reported to extract NQs. Among those organic solvents, ethanolic and methanolic extracts appear to result in a relatively purer form of NQs with limited interfering compounds. The overall protocol used to produce, purify, and characterize 1,4-NQs from plant material is illustrated in Figure 1B. Juglone, plumbagin, lawsone, shikonin, and lapachol are a few examples of 1,4-NQs found profusely in nature and are also recognized for their substantial antimicrobial potential [46]. Lim et al. [30] extracted 5-hydroxy-1,4-naphthoquinone (juglone) (A in Table 1) from Caesalpinia sappan heartwood and examined its activity against intestinal cultures, viz. Bifidobacterium bifidum, Bifidobacterium breve, Clostridium perfringens, Escherichia coli, and Lactobacillus casei. Authors had purified those bioactive molecules using different solvents, like methanol, EA, butanol, chloroform, hexane, and water. The powerful growth-inhibition activity of NQ was obtained from EA, butanol, and methanol extracts against B. bifidum and C. perfringens. Chloroform fractions resulted in moderate antimicrobial activity, whereas water and hexane fractions were ineffective against both pathogens. The scenario presented here is quite interesting and justifies the selectivity and specificity of bioactive compounds isolated from C. sappan heartwood using different solvents. Authors also screened commercially accessible compounds like 5-hydroxy-1,4-NQ, 5-hydroxy-2-methyl-1,4-NQ, 1,4-NQ, and 1,2-NQ against the above-mentioned test organisms at concentrations of 0.25, 0.5, 1, 2, and 5 mg/disk. Their analysis recommended the dose-dependent activity of 1,4-NQ, and 1,2-NQ against C. perfringens, rather than other isolates used by them. Additionally, the 1,4-NQ derivative showed superior activity against four other organisms (B. bifidum, B. breve, E. coli, and L. casei) as compared to other derivatives (5-hydroxy-1,4-NQ and 5-hydroxy-2-methyl-1,4-NQ). Results also confirmed that the antibacterial activity of 1,4-NQ was reduced significantly due to the presence of a –CH3 group at the 2nd and an –OH group at the 5th position.
Plumbagin (B in Table 1) is another widely explored secondary plant metabolite. Plumbagin is of immense interest due to its abilities to (1) generate ROS, (2) inhibit EPs, (3) accumulate drugs, and (4) cure plasmids from bacteria. Thus, due to such exceptional properties, it has been utilized as an effective antibacterial agent [47]. Periasamy et al. [31] evaluated the antibacterial activity of plumbagin (from Plumbago zeylanica) against 100 methicillin-resistant S. aureus (MRSA) strains, including MDR phenotypes. P. zeylanica root extract was obtained using ACN, DMSO, DMF, IPA, methanol and water. All fractions were used to analyze their antibacterial potential where active constituents of ACN and water showed the highest and lowest antibacterial activity respectively. Thus, Periasamy et al. [31] demonstrated that NQ constituents obtained using ACN possess promising and consistent antibacterial activity (at MIC of 4–8 μg/mL) against MRSA strains.
Similarly, Adusei et al. [43] reported a bacteriostatic effect of plumbagin (P. zeylanica) extract using EA. The authors verified its antibacterial effect at 0.5, 8, 2, and 8 μg/mL against S. aureus, E. coli, K. pneumoniae and P. aeruginosa respectively. This research group also assessed the resistance-modulating potential of antibiotics using plumbagin at its subinhibitory concentration of 4 μg/mL. This assessment demonstrated a range of MICs for ciprofloxacin (0.25 to 2 μg/mL), amoxicillin (0.25 to 256 μg/mL), ampicillin (0.25 to 256 μg/mL), and ketoconazole (no activity) against test pathogens. Their findings presented the successful resistance-modulating potential of three antibiotics, viz. ciprofloxacin, amoxicillin, and ampicillin, with plumbagin against S. aureus and E. coli. The activity of those three antibiotics was enhanced up to 2-fold when they were used in combination with plumbagin. Surprisingly, the enhancement of the 2-fold activity of ciprofloxacin was found against P. aeruginosa, whereas the activity of amoxicillin and ampicillin was reduced to 2-fold against the same isolate. For K. pneumoniae, the activity of amoxicillin and ampicillin was found to be enhanced up to 6-fold. At the same time, the efficiency of ciprofloxacin was reduced by 2-fold for K. pneumoniae [43]. The synergistic approaches are unquestionably valuable to prove the potential of plumbagin as a favorable drug. Adusei et al. [43] further reported the considerable antibiofilm activity of plumbagin (32 to 128 μg/mL) against the above-mentioned five pathogens. The antibiofilm activity of plumbagin was noticeably higher against S. aureus as compared to ciprofloxacin. Both plumbagin and ciprofloxacin displayed antibiofilm activity (>50%) against the other three pathogens (S. aureus, E. coli, and K. pneumoniae) at 128 μg/mL concentration. The combination of plumbagin with other drugs undoubtedly impede the growth and infection of biofilm-forming pathogens.
The literature suggests that plumbago species has always been a choice NQ among the ones of natural sources. Patwardhan et al. [32] documented the effectiveness of P. auriculata root extract against 23 nosocomial pathogenic strains including P. aeruginosa, P. vulgaris, E. coli, and K. pneumoniae. Solvent extracts of plumbago roots were evaluated at various concentrations from 250 to 4000 μg/mL. Plumbago extract obtained using ethanol displayed the highest antimicrobial activity as compared with other solvents. Ethanolic based extracts impeded the growth of P. vulgaris and K. pneumoniae, followed by E. coli. It is important to mention that P. aeruginosa was found to be the least affected with the same ethanolic plumbago extract.
A report documented by Kaewbumrung and Panichayupakaranant [33] explains the stability and the yield of plumbagin from P. indica roots. For the extraction procedure, authors used ethanol, EA, DCM, diethyl ether, and isopropanol. Out of those five solvents, ethanol was found to be the most suitable to result extract (11.5% w/w) having a high amount (5.79 mg/g) of the plumbagin. Further purification and elution of pure plumbagin was achieved using a silica gel column with a mixture of hexane: EA (9.2%:0.8% v/v). This step enhanced the total plumbagin content (13.26% w/w). Impurities soluble in hexane and EA were removed to result in a purified form of a derivative. The promising bactericidal activity of plumbagin extract was observed against S. aureus, S. epidermidis, and P. acnes. Like EA, petroleum ether has been the solvent of choice to extract NQs from plant sources. Vukic et al. [44] used petroleum ether along with EA to isolate NQs from Onosma visianii. Thus, the selection of appropriate solvents is important to obtain biologically active components from plant materials. From the above discussed literature, we underscore the pivotal role of solvents to extract NQs from natural sources. Attention to this concept could be helpful to enhance the overall yield of NQs from the desired biological sources. In conclusion, plumbago species are one of the preferred natural sources to isolate NQs. Among various solvents, ethanol is a virtuous choice for the extraction of bioactive compounds, followed by EA and hexane to demonstrate the assured biological activity of NQs.
Like plumbagin, another NQ—namely lawsone (C in Table 1)—has stimulated researchers to explore its medicinal potential. The foremost report on the extraction of lawsone (2-hydroxy-1,4-NQ) from P. zeylanica was accomplished by Patwardhan et al. [34]. Authors extracted lawsone via the solvents acetone, benzene, chloroform, cyclohexane, diethyl ether, ethanol, methanol, and petroleum ether. Consequently, Patwardhan and collaborators [34] focused on the biological activity, purification, and characterization of lawsone extracted using ethanol. The antibacterial potential of ethanolic root extract (ranging from 200 to 800 µg/mL) was effective against three Gram-positive cultures (S. aureus, B. cereus, and B. subtilis) and six Gram-negative cultures (E. coli, S. typhi, Enterococcus spp., K. pneumoniae, A. baumannii, and S. dysenteriae). However, the same ethanolic extract was operative against S. marcescens and Proteus mirabilis at higher concentrations (>1600 µg/mL). Patwardhan et al. [34] also examined the plasmid-curing efficiency of lawsone, which is discussed briefly in Section 5.1.
A liposoluble red-colored pigment, shikonin (D in Table 1), is usually extracted from the roots of various plants like Alkanna tinctorial, Lithospermum erythrorhizon, or Arnebia decumbens L. Shikonin is one of the unique bioactive NQs routinely extracted from the roots of L. erythrorhizon, and is popular for several biological activities.
Lee et al. [37] isolated shikonin from the roots of L. erythrorhizon and examined its antibacterial potency against seven MRSA strains. This study revealed the MIC of shikonin to be 7.8 to 31.2 μg/mL. MRSA strains were found to be more susceptible to shikonin, as compared to ampicillin and oxacillin (antibiotics of the penicillin class, cell wall attacking). Shikonin was evaluated in combination with Tris and Triton X-100 (membrane-permeabilizing agents), sodium azide and N,N′-dicyclohexylcarbodiimide (ATPase inhibitors), and peptidoglycan (derived from S. aureus). The work was supported through experiments like (1) the broth microdilution method (to analyze the susceptibility of bacteria to antibacterial agents), (2) the time–kill test (to determine the bactericidal/bacteriostatic activity of antibacterial agents over time), and (3) transmission electron microscopy (predicting the underlying mechanism of antibacterial agents affecting cell morphology). Studies have discovered the enhanced antibacterial activity of shikonin in the presence of Tris and Triton X-100 and ATPase inhibitors. Lee et al. [37] suggested that shikonin can be proposed as a natural antibiotic and even to realize the underlying mechanism responsible for antimicrobial action. Al-Mussawi [48] had also isolated shikonin from Arnebia decumbens L. and purified it using column chromatography. Researchers proved the antibacterial activity of shikonin against P. aeruginosa, E. coli, S. aureus, and K. pneumonia. Moreover, shikonin has been widely explored for anti-inflammatory, wound-healing, antithrombotic, and anticancer properties [49]. Large-scale production of any bioactive molecule is mandatory to utilize them for pharmacological purposes. When NQs are extracted from natural plant sources, the yield and purity cannot be neglected. Recently, Huang et al. [36] developed an ultrasound-assisted extraction (UAE) technique to isolate shikonin from A. euchroma. The response surface methodology (RSM) was used to design an appropriate extraction set-up for the production of shikonin. This experiment set-up can encourage researchers to extract shikonin at ease using an energy-saving approach. Authors magnificently reported around a 1.26% yield of shikonin under the ideal extraction protocol using ultrasound power at 93 W (in 87 min at 39 °C) with a liquid–solid ratio of 11:1. The same studies supplemented the antimicrobial activity of shikonin with clinical isolates (three) along with standard ones (five) at MICs of 128–1024 μg/mL. Extracts obtained from the medicinal plants find applications in the manufacturing of ointment to treat patients with burn infections. Aljanaby [50] isolated an aqueous extract having alkannin esters and shikonin from A. tinctoria and demonstrated antibacterial activity against drug-resistant bacterial strains (395) at a 300 mg/mL concentration.
Among different NQs mentioned above, lapachol (E in Table 1) has been regularly reported to have antibacterial and anticancer properties [51]. Lapachol has been derivatized to produce a pharmacologically important molecule. Zani et al. [38] had isolated lapachol from Tabebuia ochracea (Bignoniaceae family), which was further derivatized by Souza et al. [39] to produce thiosemicarbazone and semicarbazone. In an antibacterial assay, E. faecalis and S. aureus were found to be susceptible to thiosemicarbazone lapachol (0.05 μmol/mL) and semicarbazone lapachol (0.10 μmol/mL). Along with Bignoniaceae, many other families like Verbenaceae, Proteaceae, Leguminosae, Sapotaceae, Scrophulariaceae, and Malvaceae show broad scope to extract lapachol.
An interesting report by Balachandran et al. [52] documented the isolation of an antibacterially bioactive 1,4 NQ molecule from Streptomyces sp., named bluemomycin, using EA. The EA extract displayed potent antibacterial activity against both Gram-negative bacteria as well as Gram-positive bacteria.
In the view of the present scenario, it is also vital to communicate that the microorganisms used for antimicrobial assays areS. aureus, P. aeruginosa, and E. coli, which belong to the ESKAPE category. At lower MICs, most NQs of natural origin have demonstrated better activities against Gram-positive (Staphylococcus spp., Bacillus spp.) than Gram-negative bacteria. Among Gram-negative bacteria, Pseudomonas spp. and E. coli have been frequently tested to determine the antibacterial potency of NQs (Figure 2).
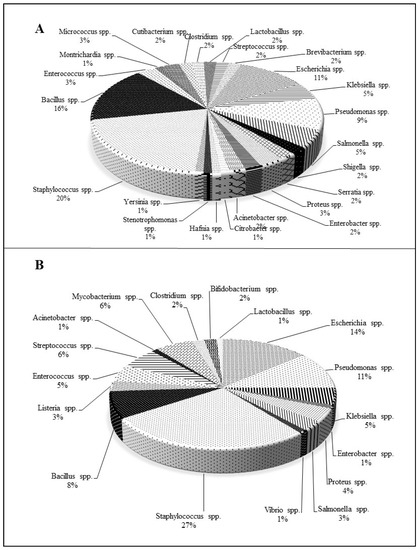
Figure 2. Percentage-wise distribution of bacterial species used to evaluate the antibacterial activity of (A): naturally derived and (B): chemically synthesized 1,4-naphthoquinones.
3.2. Chemically Synthesized 1,4-Naphthoquinone Derivatives
Just like those of a natural origin, chemically derivatized 1,4-NQs are widely investigated for antimicrobial potential. Literature suggests that most of the additions or deletions of the chemical groups have been carried out at 2nd and 3rd positions in NQ moieties. The groups preferred for additions are H, OH, CH3, Cl, Br, N, and S. Most of the chemical elements chosen for derivatization are placed from 14 to 17 in the periodic table. These atoms are typically reactive non-metals and strong oxidizing agents.
Chemical modifications in the NQ moieties have been facilitated to explore them for a wide range of biological applications. Ravichandiran et al. [53] synthesized a series of NQs containing phenylamino-phenylthio moieties and evaluated their antibacterial activity against S. aureus, Listeria monocytogenes, E. coli, P. aeruginosa, and K. pneumonia with MICs ranging between 15.6 to 500 µg/mL. Most of the synthesized NQ derivatives were able to inhibit S. aureus and E. coli. However, NQ derivatives were found to be less effective against the other three strains. The structure–activity relationship (SAR) suggested that the introduction of the thiophenol group in one of the structures can exhibit moderate antibacterial activity as compared to its ligand. This study revealed that the introduction of a substituted amide group, acid chlorides (aliphatic and aromatic), in one of the compounds shows superior antibacterial properties as compared with its parent compound. Likewise, a compound containing a 3,5-dinitro aryl moiety displayed enhanced antibacterial activity against selected pathogens. Contrary to this, synthesized compounds having a bulky moiety or electron-withdrawing groups (NO2 and methyl) showed low inhibitory actions against test cultures. This can be further justified by the fact that the bulky groups cannot enter or penetrate easily into the bacterial cell and thus, encounter challenges to fit at the target sites [53]. The synthesis of quinones substituted with N-, S-, O- and the evaluation of their antimicrobial and anticancer activities was performed by Kurban et al. [18]. For the first time, researchers documented the efficiency of benzo- and NQ derivatives to inhibit an enzyme, catalase (which is responsible for the decomposition of H2O2 into H2O and O2 to protect cells from oxidative damage).
From the literature, it is noteworthy that the bacterial species of the ESKAPE category are extensively tested to assess the efficacies of chemically synthesized NQ derivatives. NQs can impose greater effects against Gram-positive bacteria. However, it is also important to highlight the exceptional cases wherein Gram-negative strains like K. pneumoniae and E. coli are also susceptible to some of the NQ derivatives [53,54]. The mechanism of action might be influenced by the constituents of the cell wall. Also, the interaction of the functional groups of NQ derivatives at the target position is another parameter which cannot be neglected. The addition or deletion of a single chemical group (for example OH, Cl, Br, S, N, or O) to or from a specific position in a NQ structure can affect the efficacies of the NQ against targeted pathogens. Table 3 presents the antibacterial potential of chemically synthesized NQ derivatives along with their MICs.
Table 3. Antibacterial potential of chemically synthesized naphthoquinones along with their minimum inhibitory concentration (MIC).
| Type of Naphthoquinone | Antibacterial Activity Against | MIC (µg/mL) | Reference |
|---|---|---|---|
| Plumbagin | Mycobacterium tuberculosis | 4 | [56] |
| Mycobacterium smegmatis | 4 | ||
| Lawsone | Mycobacterium tuberculosis | >16 | |
| Mycobacterium smegmatis | >32 | ||
| Lawsone methyl ether | Staphylococcus aureus (MRSA) | 62.5–125 | [57] |
| Lawsone derivatives | Staphylococcus aureus (MRSA) | 32–128 & >128 | [58] |
| Staphylococcusaureus (MSSA) | 0.6 –128 & >128 | ||
| Lapachol | Staphylococcus aureus | 1.25 mM | [59] |
| Lapachol, nor lapachol | Staphylococcus aureus (MRSA) | 30–500 & >500 | [60] |
| Imidazole derivatives of 1,4-naphthoquinone | Staphylococcus aureus | 8–512 | [61] |
| Bacillus subtilis | 32–512 | ||
| Pseudomonas aeruginosa | 8–256 | ||
| Proteus vulgaris | 16–256 | ||
| Escherichia coli | 32–256 | ||
| 1,4-naphthoquinone derivatives | Staphylococcus aureus | 4–256 | [62] |
| Bacillus subtilis | 128–512 | ||
| Pseudomonas aeruginosa | 32–512 | ||
| Proteus vulgaris | 128–512 | ||
| Escherichia coli | 256–512 | ||
| Phenylamino-phenylthio derivatives of 1,4-naphthoquinone (no. of derivatives synthesized) | Staphylococcus aureus | 31.25–500 | [53] |
| Listeria monocytogenes | 62.5–500 | ||
| Pseudomonas aeruginosa | 62.5–500 | ||
| Escherichia coli | 15.6–500 | ||
| Klebsiella pneumoniae | 62.5–500 | ||
| Sulfide derivatives of 1,4-naphthoquinone | Staphylococcus aureus | 7.8–250 & >250 | [24] |
| Escherichia coli | 31.3–250 & >250 | ||
| Menadione | Staphylococcus aureus | 128 | [63] |
| Pseudomonas aeruginosa | 64 | ||
| Escherichia coli | 128 | ||
| Klebsiella pneumoniae | 128 | ||
| Arylsulfanylmethyl-[1,4]-naphthoquinone derivatives | Staphylococcus aureus | 32–256 | [64] |
| Staphylococcus epidermidis | 16–256 | ||
| Staphylococcus simulans | 32–256 | ||
| Escherichia coli | 256 (less activity) | ||
| Enterococcus faecalis | 32–256 | ||
| 1,4-naphthoquinone substituted at positions 2 and 3 | Staphylococcus aureus | 7.8–500 | [65] |
| Salmonella bongori | 125–500 | ||
| Pseudomonas aeruginosa | 125–500 | ||
| Proteus vulgaris | 250–500 | ||
| Escherichia coli | 125–500 | ||
| Klebsiella pneumoniae | 62.5–500 | ||
| Enterococcus faecalis | 125–500 | ||
| Enterobacter cloacae | 250–500 | ||
| Shikonin derivatives | Staphylococcus aureus | 1.1–45.4 | [66] |
| Bacillus subtilis | 2–50 & >50 | ||
| Escherichia coli | 3.1–50 & >50 | ||
| Pseudomonas aeruginosa | 4–50 & >50 | ||
| Naphthoquinone derivatives | Staphylococcus aureus | 16–256 | [19] |
| Pseudomonas aeruginosa | 64–256 | ||
| Escherichia coli | 64–256 | ||
| Enterococcusfaecalis | 64–256 | ||
| 2-bromo-5-hydroxy-1,4-NQ | Staphylococcus aureus | 16 | |
| Juglone | Staphylococcus aureus | 32–256 | [67] |
| Staphylococcus epidermidis | 128 | ||
| Bacillus cereus | 256 | ||
| Salmonella enterica | 256 | ||
| Listeria monocytogenes | 256 | ||
| Pseudomonas aeruginosa | 128 | ||
| Escherichia coli | 128 | ||
| Enterococcus feacalis | 256 | ||
| Vibrio alginolyticus | 256 | ||
| Nitrogen and sulfur derivatives of 1,4-naphthoquinone | Bacillus subtilis | 1.4–19.3 | [68] |
| Proteus vulgaris | 2.7–39 | ||
| Plumbagin, juglone, lawsone, menadione and their analogues | Staphylococcusaureus (MRSA) | 3.9–125 | [69] |
| Pseudomonas aeruginosa | No significant activity | ||
| Menadione | Staphylococcus aureus | 3.1 | [70] |
| Bacillus anthracis | 6.25 | ||
| Streptococcus pyogenes | 25 | ||
| Streptococcus agalactiae | 6.25 | ||
| 1,4-naphthoquinone | Staphylococcus aureus | 6.25 | |
| Bacillus anthracis | 12.5 | ||
| Streptococcus pyogenes | 50 | ||
| Streptococcus agalactiae | 12.5 | ||
| 2-hydroxy, 1,4 naphthoquinone (Lawsone) and its 2-hydroxynaphthoquinone derivatives | Staphylococcus aureus | 16–512 & >512 | [71] |
| Listeria monocytogenes | 512 & >512 | ||
| Pseudomonas aeruginosa | 512 & >512 | ||
| Escherichia coli | 512 & >512 | ||
| Enterococcus faecalis | 256–512 | ||
| Acinetobacter baumannii | 512 & >512 | ||
| Salmonella typhimurium | 512 & >512 | ||
| Plumbagin derivatives | Mycobacterium smegmatis | 13.3–30.4 | [72] |
| Mycobacterium tuberculosis | 15.6–77.4 | ||
| Nitrogen, sulfur groups substitution at 2, 3 positions of 1,4-naphthoquinone | Staphylococcus aureus | 6.25–50 & >50 | [73] |
| Escherichia coli | 6.25–50 & >50 | ||
| Klebsiella pneumoniae | 1.56–50 & >50 | ||
| 5-Hydroxy-2-methyl-1,4-NQ | Clostridium perfringens | Antibacterial activity observed however MIC values are not mentioned specifically for the test organisms | [30] |
| 1,4-naphthoquinone | Lactobacillus casei | ||
| Bifidobacterium bifidum | |||
| Bifidobacterium breve | |||
| Clostridium perfringens | |||
| Escherichia coli | |||
| 1,2-naphthoquinone | Clostridium perfringens | ||
| Bifidobacterium bifidum | |||
| Bifidobacterium breve | |||
| 5-Amino-8-Hydroxy-1,4-NQ | Staphylococcus aureus | 50 | [74] |
| 1,4-naphthoquinone | Staphylococcus aureus | 10 | |
| (L)-a-amino acid methyl ester, heteroalkyl and aryl substituted1,4-naphthoquinone derivatives | Staphylococcus aureus | 12.5–50 & >50 | [54] |
| Streptococcus faecalis | 12.5–50 & >50 | ||
| Klebsiella pneumoniae | 6.25–50 & >50 | ||
| Escherichia coli | 12.5–50 & >50 | ||
| Pseudomonas aeruginosa | 50 & >50 |
Yap et al. [55] put forth findings that demonstrated the synergistic relationship between 1,4-NQ and antibiotics like imipenem, cefuroxime, and cefotaxime against methicillin-resistant Staphylococcus aureus (MRSA). Authors suggest that 1,4-NQ could be developed into an adjuvant that will help in enhancing the function of antibiotics against drug-resistant bacteria as a part of combination therapy.
From the overall literature survey, 1,4-NQs extracted from natural sources and synthesized by chemical means propose their extraordinary candidature against pathogens. The percentage-wise distribution of bacterial species evaluated to demonstrate antibacterial potential of 1,4-NQs is depicted in Figure 2. Staphylococcus spp. belonging to the ESKAPE and MDR pathogen groups has been reported frequently for its susceptibility towards natural (~20%) and chemically synthesized (~27%) NQs. Followed by Staphylococcus spp., other Gram-positive bacteria, namely, Bacillus spp. (~16%), have been used to validate the antibacterial activity. Among Gram-negative bacteria, E. coli (~11%) and Pseudomonas spp. (~9%) seemed to be used for in vitro assays. In addition to the above-mentioned prominent bacteria, several other organisms have been utilized in experiments. Like the natural NQs, chemically synthesized NQ derivatives have generally been tested against Pseudomonas spp. and E. coli (both members of the ESKAPE and MDR pathogen groups). Organisms, viz. E. faecium, K. pneumoniae, A. baumannii, and Enterobacter spp. of ESKAPE, have been assessed for the antibacterial efficiency of NQs (Figure 2). The pragmatic analysis of 1,4-NQs, overtly showcases them as an emerging drug against the targeted pathogens.
References:
- Gaynes, R. The discovery of Penicillin—New insights after more than 75 years of clinical use. Emerg. Infect. Dis. 2017, 23, 849–853. [Google Scholar] [CrossRef]
- Horner, W.H. Streptomycin. In Biosynthesis; Springer: Berlin/Heidelberg, Germany, 1967; pp. 373–399. [Google Scholar]
- Global Pharma News and Resources. Available online: https://www.pharmiweb.com/press-release/2020-07-02/global-antibiotics-market-is-expected-to-reach-at-a-cagr-of-47-during-the-forecast-period-2019-202 (accessed on 8 September 2020).
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Tanwar, J.; Das, S.; Fatima, Z.; Hameed, S. Multidrug resistance: An emerging crisis. Interdiscip. Perspect. Infect. Dis. 2014, 2014, 541340. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua=1 (accessed on 10 November 2020).
- World Health Organization. Available online: https://www.who.int/news-room/events/detail/2020/11/18/default-calendar/world-antimicrobial-awareness-week-2020 (accessed on 16 November 2020).
- Nikaido, H. Multidrug resistance in bacteria. Annu. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef] [PubMed]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed Res. Int. 2016, 2016, 475067. [Google Scholar] [CrossRef] [PubMed]
- Sharifi-Rad, J.; Salehi, B.; Stojanović-Radić, Z.Z.; Fokou, P.V.T.; Sharifi-Rad, M.; Mahady, G.B.; Sharifi-Rad, M.; Masjedi, M.-R.; Lawal, T.O.; Ayatollahi, S.A.; et al. Medicinal plants used in the treatment of tuberculosis-ethnobotanical and ethnopharmacological approaches. Biotechnol. Adv. 2020, 44, 107629. [Google Scholar] [CrossRef] [PubMed]
- Tiwari Pandey, A.; Pandey, I.; Hachenberger, Y.; Krause, B.C.; Haidar, R.; Laux, P.; Luch, A.; Singh, M.P.; Singh, A.V. Emerging paradigm against global antimicrobial resistance via bioprospecting of mushroom into novel nanotherapeutics development. Trends Food Sci. Technol. 2020, 106, 333–344. [Google Scholar] [CrossRef]
- Tiwari Pandey, A.; Pandey, I.; Zamboni, P.; Gemmati, D.; Kanase, A.; Singh, A.V.; Singh, M.P. Traditional herbal remedies with a multifunctional therapeutic approach as an implication in COVID-19 associated co-infections. Coatings 2020, 10, 761. [Google Scholar] [CrossRef]
- Ansari, M.H.D.; Lavhale, S.; Kalunke, R.M.; Srivastava, P.L.; Pandit, V.; Gade, S.; Yadav, S.; Laux, P.; Luch, A.; Gemmati, D.; et al. Recent advances in plant nanobionics and nanobiosensors for toxicology applications. Curr. Nanosci. 2020, 16, 27–41. [Google Scholar] [CrossRef]
- Singh, V.; Kumar, V.; Kashyap, S.; Singh, A.V.; Kishore, V.; Sitti, M.; Saxena, P.S.; Srivastava, A. Graphene oxide synergistically enhances antibiotic efficacy in vancomycin-resistant Staphylococcus aureus. ACS Appl. Bio Mater. 2019, 2, 1148–1157. [Google Scholar] [CrossRef]
- Kurban, S.; Deniz, N.G.; Sayil, C.; Ozyurek, M.; Guclu, K.; Stasevych, M.; Zvarych, V.; Komarovska-Porokhnyavet, O.; Novikov, V. Synthesis, antimicrobial properties, and inhibition of catalase activity of 1,4-naphtho-and benzoquinone derivatives containing N-, S-, O-substituted. Heteroat. Chem. 2019, 2019, 1658417. [Google Scholar] [CrossRef]
- Sánchez-Calvo, J.M.; Barbero, G.R.; Guerrero-Vásquez, G.; Durán, A.G.; Macías, M.; Rodríguez-Iglesias, M.A.; Molinillo, J.M.; Macías, F.A. Synthesis, antibacterial and antifungal activities of naphthoquinone derivatives: A structure–activity relationship study. Med. Chem. Res. 2016, 25, 1274–1285. [Google Scholar] [CrossRef]
- Mathiyazhagan, K.; Kumaran, A.; Arjun, P. Isolation of natural naphthoquinones from Juglans regia and in vitro antioxidant and cytotoxic studies of naphthoquinones and the synthetic naphthofuran derivatives. Russ. J. Bioorganic Chem. 2018, 44, 346–353. [Google Scholar] [CrossRef]
- Salas, C.O.; Faúndez, M.; Morello, A.; Diego Maya, J.A.; Tapia, R. Natural and synthetic naphthoquinones active against Trypanosoma Cruzi: An initial step towards new drugs for Chagas disease. Curr. Med. Chem. 2011, 18, 144–161. [Google Scholar] [CrossRef] [PubMed]
- Jentzsch, J.; Koko, W.S.; Al Nasr, I.S.; Khan, T.A.; Schobert, R.; Ersfeld, K.; Biersack, B. New antiparasitic bis-naphthoquinone derivatives. Chem. Biodivers. 2020, 17, e1900597. [Google Scholar] [CrossRef] [PubMed]
- Lall, N.; Weiganand, O.; Hussein, A.A.; Meyer, J.J.M. Antifungal activity of naphthoquinones and triterpenes isolated from the root bark of Euclea natalensis. South Afr. J. Bot. 2006, 72, 579–583. [Google Scholar] [CrossRef]
- Wellington, K.W.; Nyoka, N.B.; McGaw, L.J. Investigation of the antibacterial and antifungal activity of thiolated naphthoquinones. Drug Dev. Res. 2019, 80, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Tandon, V.K.; Singh, R.V.; Yadav, D.B. Synthesis and evaluation of novel 1,4-naphthoquinone derivatives as antiviral, antifungal and anticancer agents. Bioorg. Med. Chem. Lett. 2004, 14, 2901–2904. [Google Scholar] [CrossRef] [PubMed]
- Schuck, D.C.; Ferreira, S.B.; Cruz, L.N.; Da Rocha, D.R.; Moraes, M.S.; Nakabashi, M.; Rosenthal, P.J.; Ferreira, V.F.; Garcia, C.R. Biological evaluation of hydroxynaphthoquinones as anti-malarials. Malar. J. 2013, 12, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Klaus, V.; Hartmann, T.; Gambini, J.; Graf, P.; Stahl, W.; Hartwig, A.; Klotz, L.-O. 1,4-Naphthoquinones as inducers of oxidative damage and stress signaling in HaCaT human keratinocytes. Arch. Biochem. Biophys. 2010, 496, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Cai, A.; Chen, G.; Xi, H.; Wu, X.; Cui, J.; Zhang, K.; Zhao, X.; Yu, J.; Wei, B.; et al. Shikonin induces mitochondria-mediated apoptosis and enhances chemotherapeutic sensitivity of gastric cancer through reactive oxygen species. Sci. Rep. 2016, 6, 38267. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shen, G.N.; Luo, Y.H.; Piao, X.J.; Jiang, X.Y.; Meng, L.Q.; Wang, Y.; Zhang, Y.; Wang, J.R.; Wang, H.; et al. Novel 1,4-naphthoquinone derivatives induce apoptosis via ROS-mediated P38/MAPK, Akt and STAT3 signaling in human hepatoma Hep3B cells. Int. J. Biochem. Cell Biol. 2018, 96, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.-Y.; Jeon, J.-H.; Jeong, E.-Y.; Lee, C.-H.; Lee, H.-S. Antimicrobial activity of 5-hydroxy-1,4-naphthoquinone isolated from Caesalpinia sappan toward intestinal bacteria. Food Chem. 2007, 100, 1254–1258. [Google Scholar] [CrossRef]
- Periasamy, H.; Iswarya, S.; Pavithra, N.; Senthilnathan, S.; Gnanamani, A. In vitro antibacterial activity of plumbagin isolated from Plumbago zeylanica L. against methicillin-resistant Staphylococcus aureus. Lett. Appl. Microbiol. 2019, 69, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, R.B.; Shinde, P.S.; Chavan, K.R.; Devale, A. Reversal of plasmid encoded antibiotic resistance from nosocomial pathogens by using Plumbago auriculata root extracts. Int. J. Curr. Microbiol. Appl. Sci. 2015, 2, 187–198. [Google Scholar]
- Kaewbumrung, S.; Panichayupakaranant, P. Antibacterial activity of plumbagin derivative-rich Plumbago indica root extracts and chemical stability. Nat. Prod. Res. 2014, 28, 835–837. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, R.B.; Dhakephalkar, P.K.; Chopade, B.A.; Dhavale, D.D.; Bhonde, R.R. Purification and characterization of an active principle, lawsone, responsible for the plasmid curing activity of Plumbago zeylanica root extracts. Front. Microbiol. 2018, 9, 2618. [Google Scholar] [CrossRef] [PubMed]
- Tekin, V.; Muftuler, F.Z.B.; Guldu, O.K.; Kilcar, A.Y.; Medine, E.I.; Yavuz, M.; Unak, P.; Timur, S. Biological affinity evaluation of Lawsonia inermis origin lawsone compound and its radioiodinated form via in vitro methods. J. Radioanal. Nucl. Chem. 2015, 303, 701–708. [Google Scholar] [CrossRef]
- Huang, X.Y.; Fu, H.L.; Tang, H.Q.; Yin, Z.Q.; Zhang, W.; Shu, G.; Yin, L.Z.; Zhao, L.; Yan, X.R.; Lin, J.C. Optimization extraction of shikonin using ultrasound-assisted response surface methodology and antibacterial studies. Evid. Based Complement Alternat. Med. 2020, 2020, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Lee, D.Y.; Kim, Y.B.; Lee, S.W.; Cha, S.W.; Park, H.W.; Kim, G.S.; Kwon, D.Y.; Lee, M.H.; Han, S.H. The mechanism underlying the antibacterial activity of shikonin against methicillin-resistant Staphylococcus aureus. Evid. Based Complement Alternat. Med. 2015, 2015, 520578. [Google Scholar] [CrossRef]
- Zani, C.L.; De Oliveira, A.B.; De Oliviera, G.G. Furanonaphthoquinones from Tabebuia Ochracea. Phytochemistry 1991, 30, 2379–2381. [Google Scholar] [CrossRef]
- Souza, M.A.; Johann, S.; Lima, L.A.R.d.S.; Campos, F.F.; Mendes, I.C.; Beraldo, H.; Souza-Fagundes, E.M.d.; Cisalpino, P.S.; Rosa, C.A.; Alves, T.M.d.A.; et al. The antimicrobial activity of lapachol and its thiosemicarbazone and semicarbazone derivatives. Mem. Inst. Oswaldo Cruz. 2013, 108, 342–351. [Google Scholar] [CrossRef]
- Widhalm, J.R.; Rhodes, D. Biosynthesis and molecular actions of specialized 1,4-naphthoquinone natural products produced by horticultural plants. Hortic. Res. 2016, 3, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.-Y.; Zhu, B.-W.; Wang, X.-D.; Qin, L.; Li, D.-M.; Miao, L.; Murata, Y. Stability of polyhydroxylated 1,4-naphthoquinone pigment recovered from spines of sea urchin Strongylocentrotus nudus. Int. J. Food Sci. Technol. 2012, 47, 1479–1486. [Google Scholar] [CrossRef]
- Carriço, M.d.P.S.B.; do Carmo Cardoso, M.F.; Da Silva, F.D.C.; Ferreira, V.F.; Lima, E.S.; Souza, J.V.B. Antifungal activity of synthetic naphthoquinones against dermatophytes and opportunistic fungi: Preliminary mechanism-of-action tests. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 26. [Google Scholar]
- Adusei, E.; Adosraku, R.K.; Oppong-Kyekyeku, J.; Amengor, C.D.; Jibira, Y. Resistance modulation action, time-kill kinetics assay, and inhibition of biofilm formation effects of plumbagin from Plumbago zeylanica Linn. J. Trop. Med. 2019, 2019, 31. [Google Scholar] [CrossRef]
- Vukic, M.D.; Vukovic, N.L.; Djelic, G.T.; Popovic, S.L.; Zaric, M.M.; Baskic, D.D.; Krstic, G.B.; Tesevic, V.V.; Kacaniova, M.M. Antibacterial and cytotoxic activities of naphthoquinone pigments from Onosma visianii Clem. EXCLI J. 2017, 16, 73–88. [Google Scholar] [PubMed]
- Petrosyan, M.; Shcherbakova, Y.; Sahakyan, N.; Vardanyan, Z.; Poladyan, A.; Popov, Y.; Trchounian, A. Alkanna orientalis (L.) Boiss. Plant isolated cultures and antimicrobial activity of their extracts: Phenomenon, dependence on different factors and effects on some membrane-associated properties of bacteria. Plant Cell Tissue Organ Cult. 2015, 122, 727–738. [Google Scholar] [CrossRef]
- Wang, J.; Cheng, Y.; Wu, R.; Jiang, D.; Bai, B.; Tan, D.; Yan, T.; Sun, X.; Zhang, Q.; Wu, Z. Antibacterial activity of juglone against Staphylococcus aureus: From apparent to proteomic. Int. J. Mol. Sci. 2016, 17, 965. [Google Scholar] [CrossRef] [PubMed]
- Padhye, S.; Dandawate, P.; Yusufi, M.; Ahmad, A.; Sarkar, F.H. Perspectives on medicinal properties of plumbagin and its analogs. Med. Res. Rev. 2012, 32, 1131–1158. [Google Scholar] [CrossRef] [PubMed]
- Al-Mussawi, A.A. Isolation and identification of shikonin from Arnebia Decumbens L. and its antibacterial activity. Res. J. Appl. Sci. 2010, 6, 1452–1456. [Google Scholar]
- Andújar, I.; Ríos, J.L.; Giner, R.M.; Recio, M.C. Pharmacological properties of shikonin-a review of literature since 2002. Planta Med. 2013, 79, 1685–1697. [Google Scholar] [CrossRef]
- Aljanaby, A.J. Antibacterial Activity of an aqueous extracts of Alkanna tinctoria roots against drug resistant aerobic pathogenic bacteria isolated from patients with burns infections. Russ. Open Med. J. 2018, 7, e0104. [Google Scholar] [CrossRef]
- De Almeida, E.R. Preclinical and clinical studies of lapachol and beta-lapachone. Open Nat. Prod. J. 2009, 2, 42–47. [Google Scholar] [CrossRef]
- Balachandran, C.; Al-Dhabi, N.A.; Duraipandiyan, V.; Ignacimuthu, S. Bluemomycin, a new naphthoquinone derivative from Streptomyces sp. with antimicrobial and cytotoxic properties. Biotechnol. Lett. 2021, 43, 1–4. [Google Scholar]
- Ravichandiran, P.; Sheet, S.; Premnath, D.; Kim, A.R.; Yoo, D.J. 1,4-Naphthoquinone analogues: Potent antibacterial agents and mode of action evaluation. Molecules 2019, 24, 1437. [Google Scholar] [CrossRef] [PubMed]
- Tandon, V.K.; Yadav, D.B.; Singh, R.V.; Chaturvedi, A.K.; Shukla, P.K. Synthesis and biological evaluation of novel (L)-α-amino acid methyl ester, heteroalkyl, and aryl substituted 1,4-naphthoquinone derivatives as antifungal and antibacterial agents. Bioorg. Med. Chem. Lett. 2005, 15, 5324–5328. [Google Scholar] [CrossRef] [PubMed]
This entry is adapted from the peer-reviewed paper 10.3390/coatings11040434
This entry is offline, you can click here to edit this entry!
