The survival of insects depends on their ability to detect molecules present in their environment. Odorant-binding proteins (OBPs) form a family of proteins involved in chemoreception. While OBPs were initially found in olfactory appendages, recently these proteins were discovered in other chemosensory and non-chemosensory organs. OBPs can bind, solubilize and transport hydrophobic stimuli to chemoreceptors across the aqueous sensilla lymph. In addition to this broadly accepted “transporter role”, OBPs can also buffer sudden changes in odorant levels and are involved in hygro-reception. The physiological roles of OBPs expressed in other body tissues, such as mouthparts, pheromone glands, reproductive organs, digestive tract and venom glands, remain to be investigated.
- insect
- olfaction
- taste
- chemosensory functions
- non-chemosensory functions
- odorant-protein-binding assay
- Drosophila melanogaster
1. Introduction
Chemoperception allows organisms to detect nutritive food and avoid toxic compounds. Moreover, chemoperception is necessary for animals to identify suitable ecological niches and mating partners. Chemoreception is mediated by chemosensory receptors that interact with a variety of semio-chemicals, (odorants, pheromones and sapid molecules), allowing their detection and eliciting an adapted behaviour. In insects, the dendrites of the sensory neurons found in olfactory and gustatory sensilla are bathed in an aqueous phase called the sensillar lymph. Therefore, volatile and non-volatile chemical compounds contacting sensory organs should be solubilized and transported across the internal aqueous phase before reaching the sensory receptors. These carrier mechanisms, called “peri-receptor events” [1], involve several families of proteins, including odorant-binding proteins (OBPs). OBPs are small soluble proteins found in high concentration in both the nasal mucus of vertebrates and the chemo-sensilla lymph of insects [2][3][4][5][6][7]. OBPs were initially discovered during the early 1980s in parallel by two research groups working on the cow [8][9][10] and on the giant moth Antheraea Polyphemus [11]. A large number of DNA sequences encoding OBPs were later identified in several vertebrate species, including rat [12], pig [13][14], xenopus [15], and human [16][17]. OBPs were also detected in more than one hundred insect species, such as the silk moth Bombyx mori [18][19], the gypsy moth Lymntria dispar [20], the turnip moth Agrotis segetum [21][22], the stemborer Sesamia nonagrioides [23], the cotton bollworm Helicoverpa armigera and the oriental tobacco budworm Helicoverpa assulta [24].
2. Expression Pattern of Insect OBPs
2.1. Number of OBP-Coding Genes in Insects
The number of OBP-coding genes is highly variable between insect species, ranging between 13 in some ant species [25] to >100 in several mosquitoes [26] (Table 1).
Table 1. Numbers of annotated odorant-binding proteins (OBP) genes in different insects [27][28][29].
| Species | OBP Gene Number | |
|---|---|---|
 |
D. melanogaster | 52 |
| D. simulans | 52 | |
| D. sechellia | 51 | |
| D. yakuba | 55 | |
| D. erecta | 50 | |
| D. ananassae | 50 | |
| D. pseudoobscura | 45 | |
| D. persimilis | 45 | |
| D. willistoni | 62 | |
| D. mojavensis | 43 | |
| D. virilis | 41 | |
| D. grimshaw | 46 | |
 |
Anophele gambiae | 69 |
| Culex quinquefasciatus | 109 | |
| Aedes aegypti | 111 | |
 |
Tribolium castaneum | 49 |
 |
Apis mellifera | 21 |
 |
Blatella germanica | 109 |
 |
Solenopsis invicta | 18 |
 |
Bombyx mori | 44 |
2.2. Evolution of OBP Genes
Exhaustive comparative genomic analysis of OBPs gene families in 20 Arthropoda species revealed a highly dynamic evolution, with a high number of gains and losses of genes. The number of OBP members is variable and diverse across Arthropoda species, exhibiting a wide range of gene lengths and encoding different cysteine profiles. Interestingly, two OBP members (OBP73a and OBP59a) have clear orthological relationships not only in the 12 Drosophila genomes but also in almost all insect species (except in Hymenoptera). Studies in the organization in chromosome clusters of OBP genes showed that this gene family is significantly clustered across the Drosophila evolution. This conservation across ∼400 myr of evolution suggests the existence of some functional constraints maintaining the clusters [30]. Other reports revealed that OBPs were only present in the Hexapoda (insects), and absent in other arthropod subphyla including the non-hexapod pan-crustaceans, chelicerates and myriapods. Moreover, OBP genes were detected in ancestral hexapods, such as Archaeognatha, Zygentoma, and Phasmatodea. However, the origin of OBP genes is still unknown and needs further investigation [31][32].
2.3. Tissue Expression and Cellular Localization of OBPs
Insect OBPs were originally identified in olfactory sensilla (Vogt and Riddiford, 1981) using immuno-electron microscopy, which enable the determination of their expression patterns in the different antennal sensilla types (trichoid, basiconic and coeloconic). A comparative study conducted on three moth species, the saturniid Antheraea polyphemus, the bombycid Bombyx mori, and the noctuid Autographa gamma, detected PBPs in trichoid sensilla, particularly in the extracellular sensillum lymph of the hair lumen and in the sensillum-lymph cavities. Moth PBPs were also detected in secretory organelles of the trichogen and tormogen cells, supporting the hypothesis that these cells can produce and secrete PBPs into the sensillar lymph [33].
Recently, Larter et al. focused on the ten OBPs most abundantly expressed in the Drosophila antenna. They used in situ hybridization to map their spatial distribution in the different morphological sensilla classes (Figure 1a). The expression profiles of these antennal OBPs were more precisely investigated in the basiconic sensilla subtypes using double-labelling with OBP and OR markers. The expression patterns of distinct OBP subsets in different basiconic sensilla were identified. The map reveals that ab8 and ab9 basiconic sensilla express only one abundant OBP (OBP28a), while others co-express different OBPs. Moreover, some functionally distinct basiconic sensilla contain the same subset of abundant OBPs (Figure 1b) [34]. Drosphila olfactory and gustatory sensilla house three accessory cells that surround the cell body of the sensory neurons: the thecogen, tormogen and trichogen cells which are involved in insect sensilla morphogenesis and in OBP expression in the lymph. Conversely, non-neuronal cells expressing OBPs in antennal sensilla were identified using markers labelling each accessory cell. This study revealed that OBPs can be either expressed in tormogen cells or in thecogen cells. The only exception was OBP28a, which is simultaneously expressed in both types of accessory cell [34].

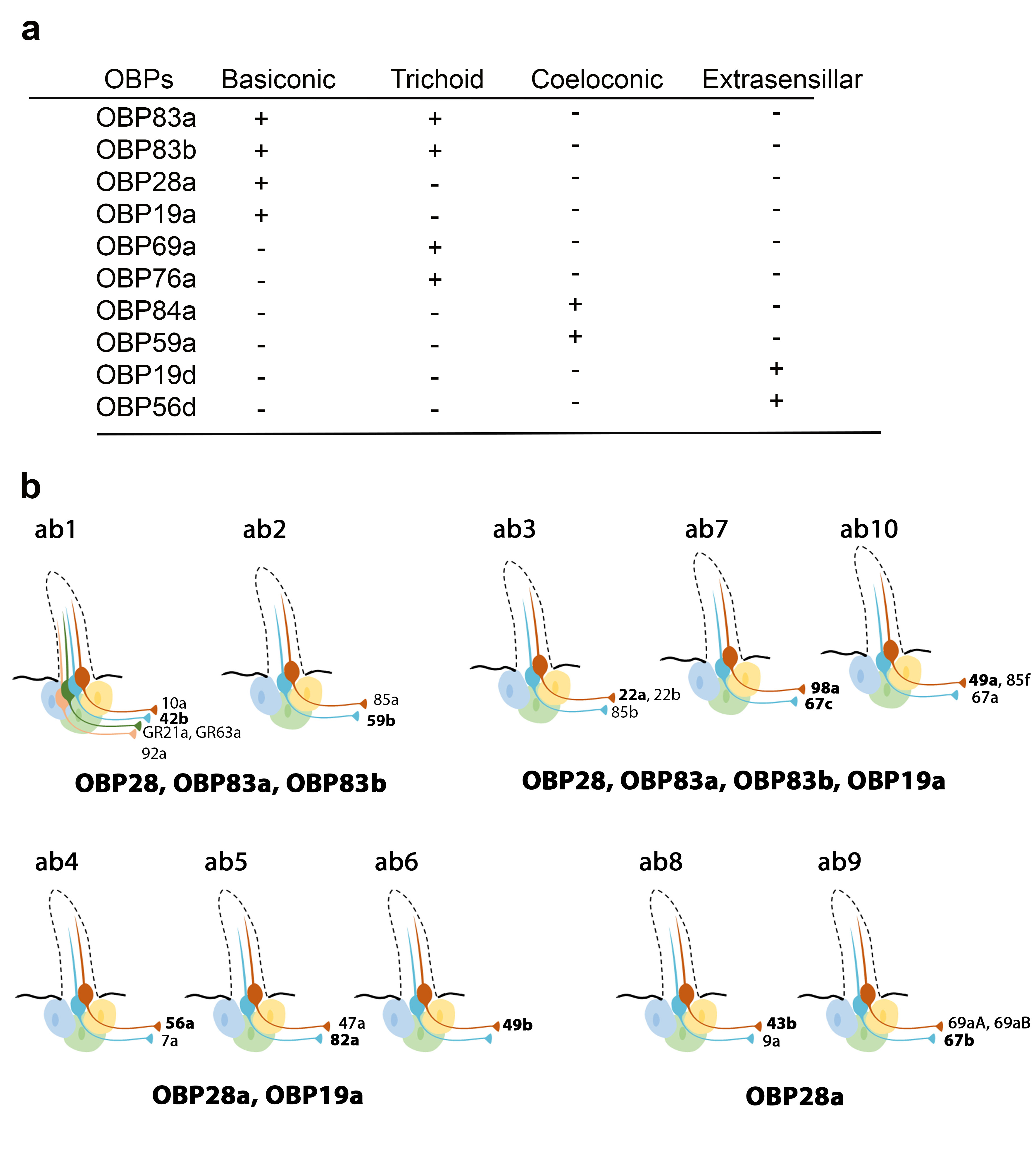
Figure 1. Expression patterns of the most abundant OBPs in Drosophila antennae. (a) Summary of the OBP expression patterns in the three types of antennal sensilla. OBP19d and OBP56g are expressed in epidermal cells. (b) Distribution of highly abundant OBPs expressed in Drosophila antennal basiconic sensilla. Olfactory receptor (OR) genes expressed in each olfactory receptor neuron class are indicated (all the OR genes except for GR21a and GR63a). The OR markers used for in situ hybridization for each sensilla are shown in bold [28][34].
The spatial and temporal expression patterns of insect OBPs have been reported in several studies, revealing that OBPs are expressed in both olfactory and taste appendages or in either chemosensory system. Gustatory OBPs have been less commonly studied than those expressed in olfactory tissues. For example, OBP57d and OBP57e are expressed in specific leg sensilla of different Drosophila species [35], while OBP49a and OBP19b are expressed in thecogen cells of D. melanogaster labellar sensilla [36][37]. The OBP19b protein was only detected in small and intermediate proboscis sensilla [37]. Moreover, OBP19d is not only expressed in olfactory appendages of D. melanogaster (antenna and maxillary palps) but also in adult gustatory organs (labellar bristles and pegs, legs, wings and in ventral and dorsal cibarial sense organs (VSCO)) [38][39] (Figure 2a,b). Two Helicoverpa armigera OBPs and one Plutella xylostella OBP were detected in the mouthparts [40], while OBP57e, OBP56g, OBP28a2 and OBP49a were identified in the legs of the oriental fruit fly Bactrocera dorsalis [41]. In Culex pipiens quinquefasciatus adults, several OBPs were exclusively identified in olfactory tissues, while others (OBP10, OBP17, OBP18, OBP22, OBP25) were identified in taste appendages (proboscis and legs) [42]. Similarly, taste-specific OBPs were identified in the labellum and tarsi in Aedes aegypti and in the red flour beetle Tribolium castaneum [43][44]. In the desert locust Schistocerca gregaria, a subset of the antennal OBP repertoire is also expressed in the maxillary and the labial palps [45]. Moreover, seven genes expressed in the labellum and tarsus of the fleshfly Boettcherisca peregrina were identified and show sequence similarity to insect OBP genes. Homologues of these gene products were detected in D. melanogaster taste tissues [46]. In the legs of the two mosquito species Anopheles gambiae and Anopheles arabiensis, the identified OBP (agCP1564) shows high similarity to Drosophila OBP57e, which is specifically expressed in the tarsi [47][48] (Figure 2a). Notably, 6 OBPs (OBP1, OBP2, OBP3, OBP4, OBP7 and OBP8) were found in the antenna and legs of the onion fly Delia antiqua. Homology studies identified their D. melanogaster homologues (OBP19d, OBP83a, OBP83b, OBP56h, OBP76a, OBP69a, respectively). Unlike D. antiqua, D. melanogaster homologues are only expressed in the fly antenna except for OBP19d, which is also expressed in Drosophila tarsi, and OBP56h, which is also expressed in Drosophila proboscis [49] (Figure 2a,b). Other studies have reported the expression of OBPs in insect legs: OBP7 in B. dorsalis [50], OBP10 in Clostera restitura [51] and OBP4, OBP6, OBP7, OBP8 in Adelphocoris lineolatus [52]. Similarly, OBPs are expressed in the legs and wings of three species of social hymenopterans (Polistes dominulus, Vespa crabro, Apis mellifera) [53]. Notably, three OBPs were identified in the anterior margin of the wings of D. melanogaster [39][47] (Figure 2a). The differences in OBP expression between tarsi, labellum and wings might be explained by the distinct roles of OBPs in food detection and intake.
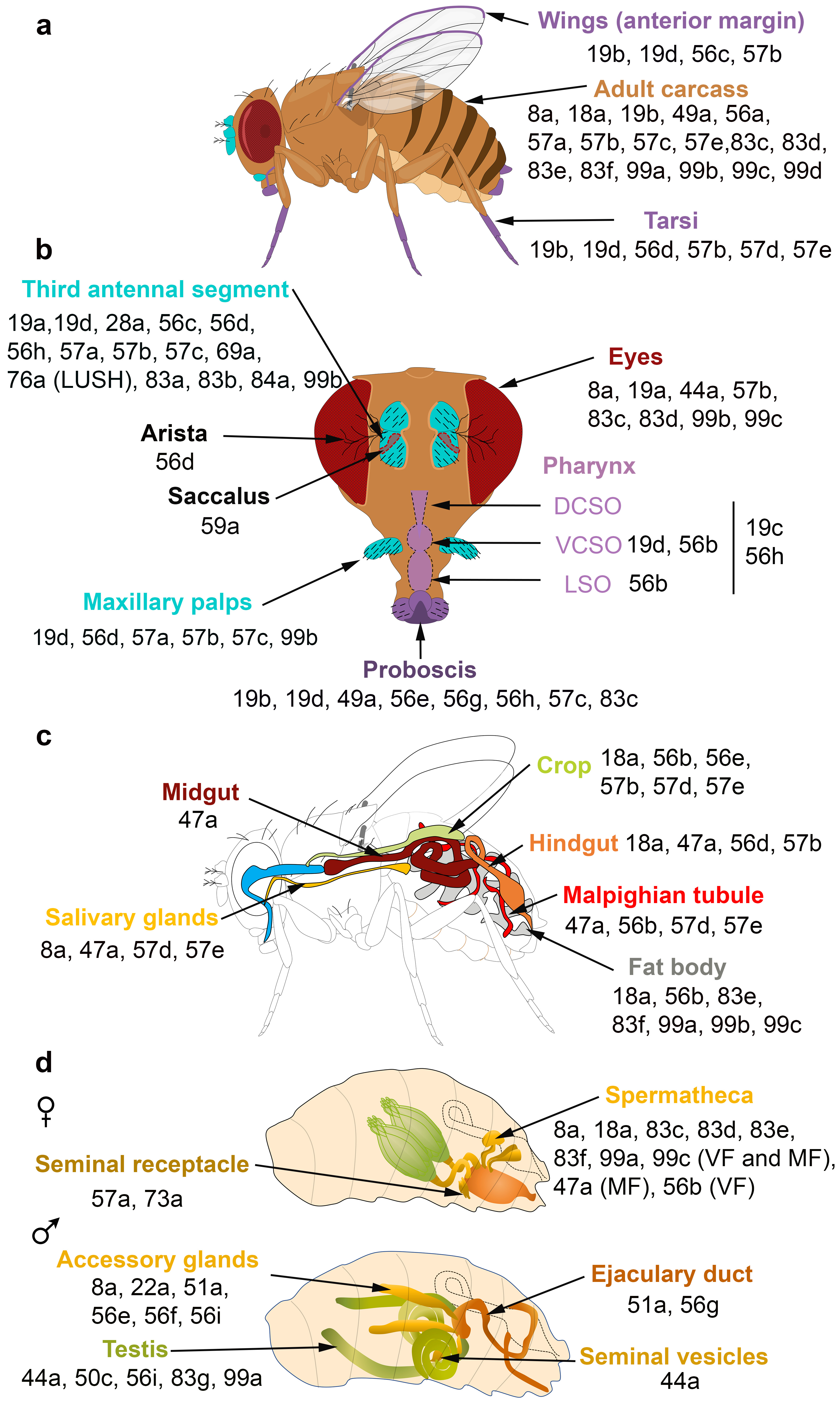
Figure 2. OBPs expressed in the olfactory, gustatory, digestive and reproductive organs of D. melanogaster. (a,b) Adult taste organs (purple) consist of the proboscis, the internal structure in the pharynx (the labral sense organ (LSO), the ventral and dorsal cibarial sense organs (VCSO and DCSO)), the leg tarsi, the anterior margin of the wings and the female genitalia. Adult olfactory organs (turquoise) consist of the antenna and the maxillary palps. OBPs expressed in each organ are indicated by their number. OBPs are found in chemosensory and non-chemosensory organs. (c) Expression patterns of OBPs in the adult digestive tract. (d) Expression patterns of OBPs in the female and male reproductive organs. The expression of OBPs in female spermatheca depends on the sexual state. Some OBPs are expressed in virgin female (VF) and mated female (MF) spermatheca, while the expression of some OBPs is mating-dependent [36][37][38][47][54][55][56][57][58][59].
Proteomic and transcriptional studies confirmed the expression of a subset of insect OBPs in non-sensory organs. In the honeybee, 9 of the 21 OBPs predicted by the genomic sequence were detected in the mandibular glands [60]. OBPs can be expressed in female and male reproductive organs. In D. melanogaster, six OBPs (especially the abundant OBP56f and OBP56g) were detected among the seminal fluid proteins transferred to females during copulation, and three of these OBPs were found in the seminal receptacle [55][61][62][63]. Similarly, OBP10 is highly abundant in the seminal fluid of the two Lepidopteran species Helicoverpa armigera and H. assulta [24]. The OBPs present in seminal fluid could be carriers of oviposition deterrents. In addition, OBP22 of the mosquito A. aegypti [64][65], OBP9 of A. mellifera [66] and two OBPs of Tribolium castaneum [67] are also present in sperm. Proteomic analysis revealed OBP expression in mosquito ovaries and eggshell [68][69][70]. RNAseq analyses and RT-PCR data also revealed the presence of OBPs in the ovaries of the stemborer Sesamia nonagrioides [23]. We can hypothesize that their accumulation in the ovaries is involved in oocyte maturation. These OBPs might also bind chemo-attractant molecules, resulting in sperm attraction. Moreover, in the oriental fruit fly B. dorsalis, OBP44a, OBP49a, and OBP56g are highly expressed in the male testis and OBP19c is highly expressed in the female ovary [41]. Examination of the FlyAtlas expression database reveals that OBP44a, OBP50c, OBP56i, OBP83g, and OBP99a are expressed in D. melanogaster male testis (Figure 2d). Among the 32 OBP genes annotated in the Hessian fly Mayetiola destructor, 24 and 25 of them were found to be expressed in female and male terminal abdomens, respectively. Only OBP31 (in female) and OBP11, OBP24 and OBP32 (in male) showed relatively higher expression levels in the terminal abdomen than in the antennae [71]. Moreover, four OBPs (OBP1, OBP4, OBP8, OBP10) were identified in the B. dorsalis abdomen, which houses the reproductive organs. These OBPs share high sequence homology with their D. melanogaster analogues (OBP8, OBP56d, OBP83ef and OBP99c, respectively). D. melanogaster analogues are also expressed in different reproductive organs present in the abdomen (Figure 2d) [50]. In Culex quinquefasciatus and Anopheles funestus, OBP expression was also detected in the abdomen [42][72]. Moreover, OBP22a, OBP51a, OBP56e, OBP56f, OBP56i are highly expressed in D. melanogaster male accessory glands (Figure 2d). All these data suggest that OBPs may (i) serve to bring odorants or pheromones next to the odorant receptors present in the female reproductive tract or (ii) carry male-specific molecules into female tissue to elicit a behavioral response. It is not yet known whether these OBPs are related to fertility and fecundity features.
The FlyAtlas expression database reveals that 6 OBPs are expressed in D. melanogaster eyes (Figure 2b). Similarly, several OBPs were identified in the eyes of the lepidopteran H. armigera [40]. Together with other proteins, these OBPs may be implicated in the complex mechanism of vision, specifically in the generation, transport and recycling of visual pigments.
In D. melanogaster, some OBPs are expressed in both larva and adults, while others are only expressed in adults (Figure 3). The majority of OBPs expressed in larva show similar expression patterns in adult tissues (Figure 2 and Figure 3).
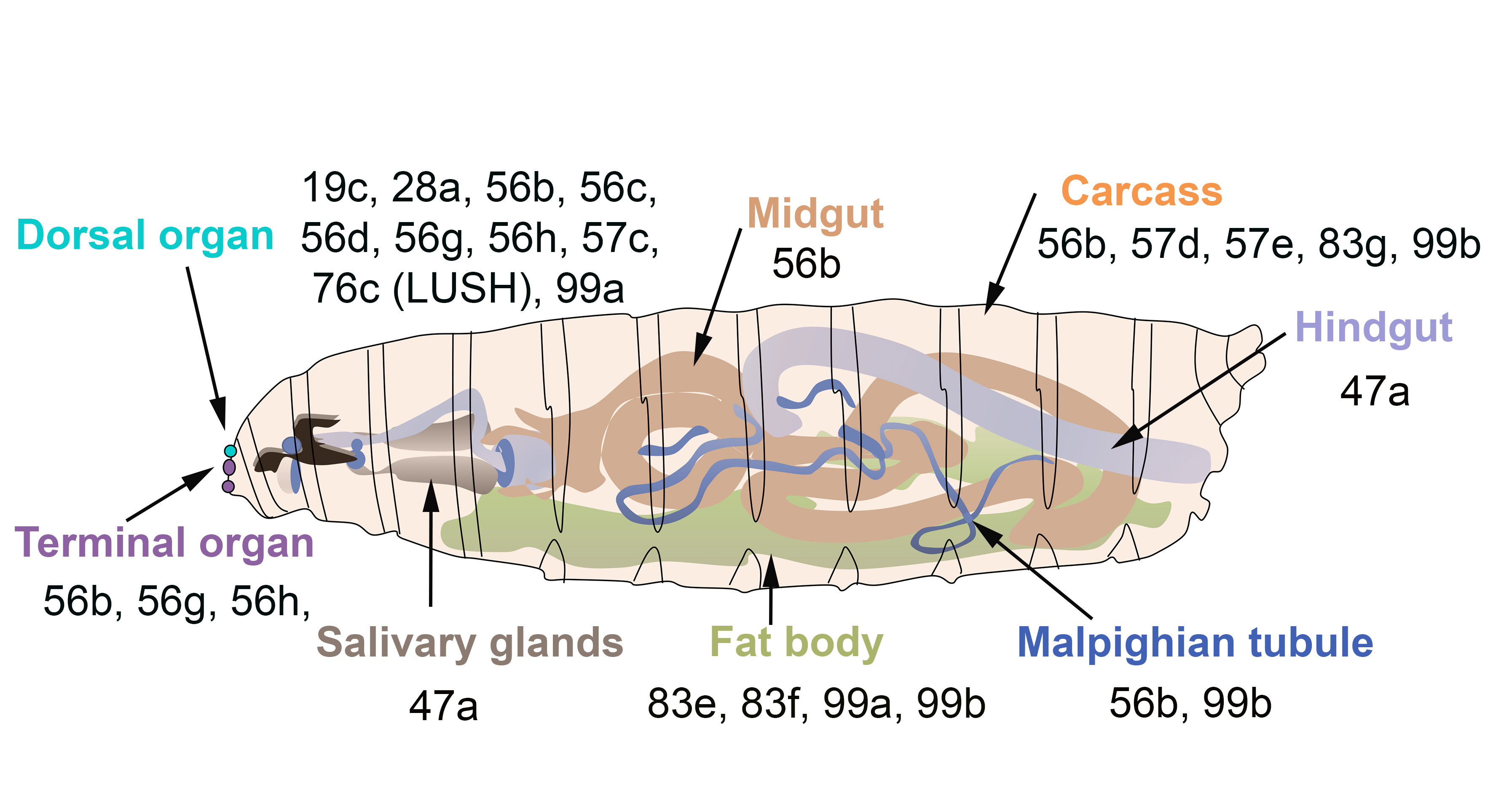
Figure 3. OBPs expressed in the olfactory gustatory and digestive organs of D. melanogaster larvae. The larval dorsal organ (turquoise) detects volatile odorants, while the larval terminal organ (purple) detects both soluble and volatile chemicals.
Surprisingly, several OBPs are expressed in the venom glands of the parasitic wasps Leptopilina heterotoma and Pteromalus puparum and of the honeybee A. mellifera [73][74][75]. Few studies have reported the expression of OBPs in the insect digestive tract. For instance, PregOBP56a was detected in the oral disk of the blowfly Phormia regina [76], while OBP56d was identified in the hindgut of D. melanogaster flies [54]. Fluorescent binding assays revealed that PregOBP56a binds palmitic, stearic, oleic, and linoleic acids. These data indicate that PregOBP56a might solubilize and deliver fatty acids to the midgut during feeding [76]. Similarly, the midgut of Rhodnius prolixus also expresses OBPs [77]. Other studies have shown that the expression of OBPs can be altered depending on the insect’s diet. Indeed, the expression of one OBP of female Culex nigripalpus increased in the midgut, thorax and abdomen after a bloodmeal, suggesting a possible role in blood feeding [78][79]. Moreover, a diet change in Anoplophora glabripennis can affect gut-expressed OBPs together with other genes implicated in digestion, detoxification and nutrient acquisition. The feeding of A. gabripennis larvae in a host with documented resistance (Populus tomentosa) induced the downregulation of 5 OBP genes. It is not known whether alteration of the gut OBP gene expression is directly linked to the resistance of A. gabripennis to the Populus tomentosa plant [80]. Moreover, bacterial symbionts increase the gut expression of tsetse’s OBP6 and of OBP28a in D. melanogaster [81]. The roles of these OBPs are discussed in a later section of this review. Notably, nineteen B. dorsalis OBPs and seven D. melanogaster OBPs are highly expressed in the fat body [41] (Figure 2c), although their roles in the fat body remain unclear. It is important to acknowledge that the expression of OBPs in specific organs does not represent a proof of function. Further physiological studies are needed to fully investigate the role(s) of OBPs in the different parts of the insect body. In the absence of selection against OBPs expression, some OBPs still become expressed despite having no obvious function. This phenomenon could lead to rapid evolution of novel functions.
3. Biochemical Properties of OBPs
The structure and binding properties of Insect OBPs were investigated. Insect OBPs are small soluble proteins classified based on the number of amino acid residues found in their primary structure: long-chain OBPs (~160 residues), medium-chain OBPs (~120 residues) and short-chain OBPs (~100 residues). The similarity between the amino acid sequences of OBPs from the same species is low (<10% identity). The protein sequences of insect OBPs include highly conserved cysteines with a specific number of amino acid residues (AAs) between them [82,83].
The three-dimensional structure of classic OBPs consists of a six α-helical domain forming a hydrophobic cavity [94] (Figure 4). The structural stability of insect OBPs depends on the presence of three interlocked disulphide bridges linking conserved cys- teines [95,96,97]. Although the AA sequences of insect OBPs are highly divergent between and within species, the structure of insect OBPs is highly conserved. To date, crystal or NMR structures of more than 20 OBPs or PBPs from species belonging to different insect orders have been solved and are available in Entrez’s 3-D structure database at NCBI, ac- companied by more than 20 detailed papers [94,98,99,100,101,102,103,104,105]. OBP structures have been solved in ligand-free (apo) or ligand-bound states, allowing researchers to study the interaction of the binding cavity with pheromones or with general odorants (Figure 4).
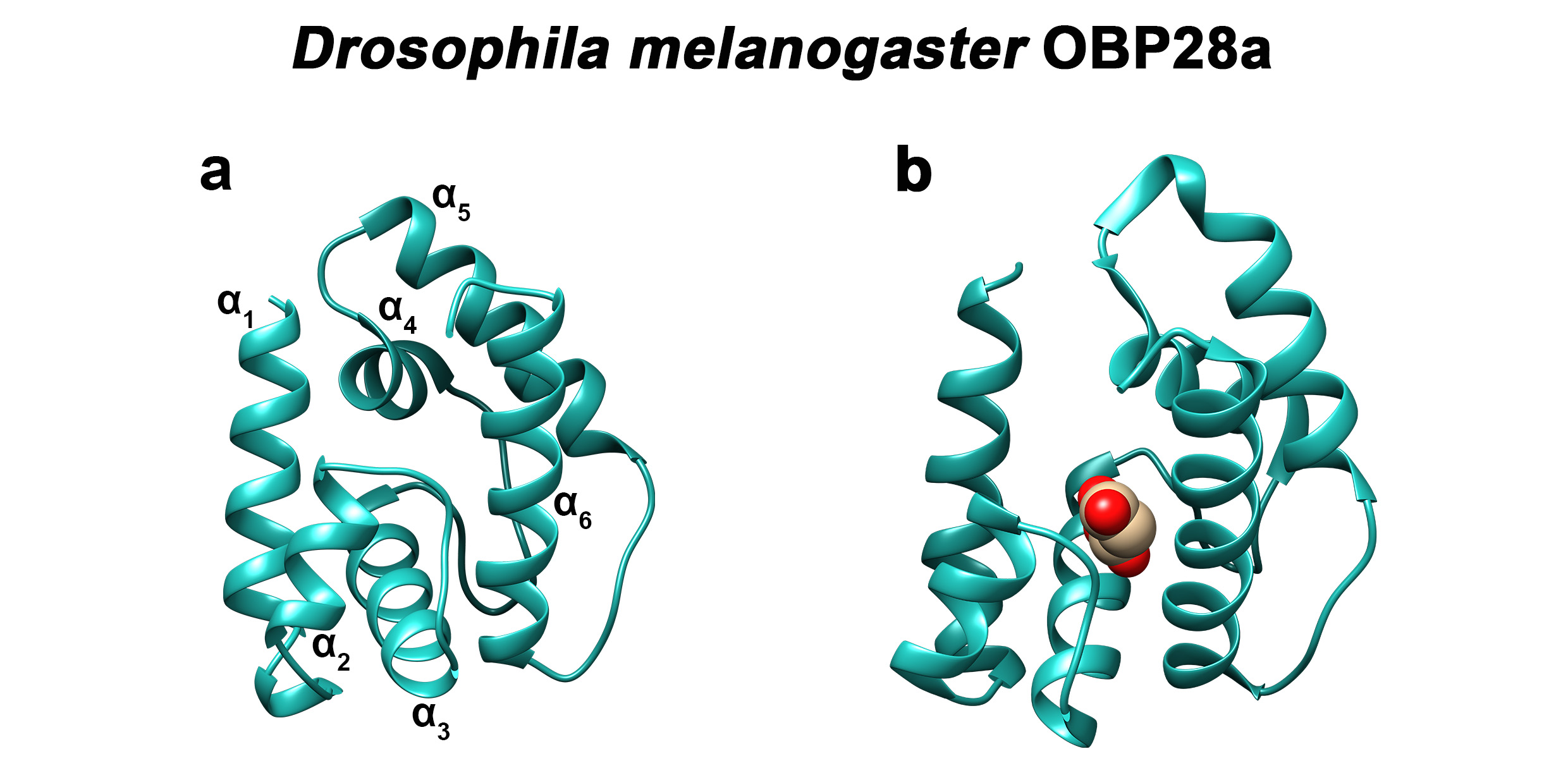
Figure 4. Three-dimensional structure of D. melanogaster OBP28a. (a) Apo- and (b) bound-three-dimensional structures of the fruit fly (Drosophila melanogaster) OBP28a (PDB: 6QQ4). α1 to α6 indicate the six α-helices found in the structure of classical insect OBPs. Drosophila OBP28a is in complex with penta-ethylene glycol [82].
The potential ligands of insect OBPs have been identified using different techniques. These studies revealed that OBPs can bind different families of chemical compounds. The physiological role of OBPs in the perception of the identified ligands was investigated in further studies using behavioural assays and electrophysiology.
4. Diverse Chemosensory Functions of OBPs
Since the discovery of OBPs, several hypotheses and models have been proposed concerning their roles in chemoreception [125]. Later, studies using structural analyses, in vitro binding, behavioural assays and electrophysiological recordings revealed unsuspected roles of insect OBPs (see Table 2).
|
OBP |
Role |
Publication |
|
OBP76a (LUSH) |
Solubilization, transport and interaction with SNMP1 |
|
|
OBP69a |
Implication in cVA response, role remains unclear |
[140] |
|
OBP28a |
Modulation of olfactory sensitivity
|
|
|
OBP59a |
Humidity detection |
[57] |
|
OBP57d and OBP57e |
Modulation of oviposition site preference to C6-C9 acids in D. melanogaster and D. sechellia Specialization of D. sechellia to its host plant (Tahitian Noni) |
[151]
[152] |
|
OBP49a |
Suppression of the appetence for sweet compounds through the perception of bitter chemicals |
[36] |
|
OBP56h |
Modulation of mating behaviour by alteration of cuticular hydrocarbon profiles in males |
[142] |
|
OBP19b |
Detection of peculiar amino acids |
[37] |
5. Conclusions
(1) Studies conducted in the last 40 years have provided information regarding the different roles of insect OBPs. OBPs were believed to be only expressed in olfactory organs and to be strictly involved in chemoreception mechanisms. However, an increasing number of reports has revealed that OBPs are expressed in most organs of the insect body and have non-conventional roles, including in taste, immunity response and humidity detection.
(2) We present the latest discoveries made on the structural and binding properties of insect OBPs. We focus on the properties of insect OBPs and, more specifically, on their tissue and cellular expression. We also present the varied functional roles, both classical and some non-conventional, of currently known OBPs.
(3) Yet there is massive amount of information on OBP functions that we ignore and needs to be investigated. These studies will pave the way to different technological applications in environmental, food quality and medical fields.
This entry is adapted from the peer-reviewed paper 10.3390/biom11040509
References
- Getchell, T.V.; Margolis, F.L.; Getchell, M.L. Perireceptor and receptor events in vertebrate olfaction. Prog. Neurobiol. 1984, 23, 317–345.
- Leal, W.S. Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu. Rev. Entomol. 2013, 58, 373–391.
- Pelosi, P.; Zhou, J.-J.; Ban, L.P.; Calvello, M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 2006, 63, 1658–1676.
- Pelosi, P.; Maida, R. Odorant-binding proteins in insects. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1995, 111, 503–514.
- Steinbrecht, R.A. Odorant-Binding Proteins: Expression and Function. Ann. N. Y. Acad. Sci. 1998, 855, 323–332.
- Tegonia, M.; Pelosi, P.; Vincenta, F.; Spinellia, S.; Campanacci, V.; Grollic, S.; Ramonic, R.; Cambillaua, C. Mammalian odorant binding proteins. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzym. 2000, 1482, 229–240.
- Vogt, R. Molecular Basis of Pheromone Detection in Insects. Compr. Mol. Insect Sci. 2005, 3, 753–803.
- Pelosi, P.; Baldaccini, N.E.; Pisanelli, A.M. Identification of a specific olfactory receptor for 2-isobutyl-3-methoxypyrazine. Biochem. J. 1982, 201, 245–248.
- Pelosi, P.; Pisanelli, A.M.; Baldaccini, N.E.; Gagliardo, A. Binding of [3H]-2-isobutyl-3-methoxypyrazine to cow olfactory mucosa. Chem. Senses 1981, 6, 77–85.
- Pevsner, J.; Hou, V.; Snowman, A.M.; Snyder, S.H. Odorant-binding protein. Characterization of ligand binding. J. Biol. Chem. 1990, 265, 6118–6125.
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nat. Cell Biol. 1981, 293, 161–163.
- Pevsner, J.; Sklar, P.B.; Snyder, S.H. Odorant-binding protein: Localization to nasal glands and secretions. Proc. Natl. Acad. Sci. USA 1986, 83, 4942–4946.
- Vincent, F.; Spinelli, S.; Ramoni, R.; Grolli, S.; Pelosi, P.; Cambillau, C.; Tegoni, M. Complexes of porcine odorant binding protein with odorant molecules belonging to different chemical classes. J. Mol. Biol. 2000, 300, 127–139.
- Spinelli, S.; Ramoni, R.; Grolli, S.; Bonicel, J.; Cambillau, C.; Tegoni, M. The Structure of the Monomeric Porcine Odorant Binding Protein Sheds Light on the Domain Swapping Mechanism. Biochemistry 1998, 37, 7913–7918.
- Millery, J.; Briand, L.; Bézirard, V.; Blon, F.; Fenech, C.; Richard-Parpaillon, L.; Quennedey, B.; Gascuel, J.; Pernollet, J.-C. Specific expression of olfactory binding protein in the aerial olfactory cavity of adult and developing Xenopus. Eur. J. Neurosci. 2005, 22, 1389–1399.
- Lacazette, E.; Gachon, A.-M.; Pitiot, G. A novel human odorant-binding protein gene family resulting from genomic duplicons at 9q34: Differential expression in the oral and genital spheres. Hum. Mol. Genet. 2000, 9, 289–301.
- Briand, L.; Eloit, C.; Nespoulous, C.; Bézirard, V.; Huet, J.-C.; Henry, C.; Blon, F.; Trotier, D.; Pernollet, J.-C. Evidence of an Odorant-Binding Protein in the Human Olfactory Mucus: Location, Structural Characterization, and Odorant-Binding Properties. Biochemistry 2002, 41, 7241–7252.
- Maida, R.; Pelosi, P. Identification and partial purification of a pheromone-binding protein in Bombyx mori. Ital. J. Biochem. 1989, 38, 211A–213A.
- Shiota, Y.; Sakurai, T.; Daimon, T.; Mitsuno, H.; Fujii, T.; Matsuyama, S.; Sezutsu, H.; Ishikawa, Y.; Kanzaki, R. In vivo functional characterisation of pheromone binding protein-1 in the silkmoth, Bombyx mori. Sci. Rep. 2018, 8, 1–8.
- Vogt, R.; Kohne, A.; Dubnau, J.; Prestwich, G. Expression of pheromone binding proteins during antennal development in the gypsy moth Lymantria dispar. J. Neurosci. 1989, 9, 3332–3346.
- Prestwich, G.D.; Du, G.; Laforest, S. How is Pheromone Specificity Encoded in Proteins? Chem. Senses 1995, 20, 461–469.
- Strandh, M.; Johansson, T.; Löfstedt, C. Global transcriptional analysis of pheromone biosynthesis-related genes in the female turnip moth, Agrotis segetum (Noctuidae) using a custom-made cDNA microarray. Insect Biochem. Mol. Biol. 2009, 39, 484–489.
- Glaser, N.; Gallot, A.; Legeai, F.; Montagné, N.; Poivet, E.; Harry, M.; Calatayud, P.-A.; Jacquin-Joly, E. Candidate Chemosensory Genes in the Stemborer Sesamia nonagrioides. Int. J. Biol. Sci. 2013, 9, 481–495.
- Sun, Y.-L.; Huang, L.-Q.; Pelosi, P.; Wang, C.-Z. Expression in Antennae and Reproductive Organs Suggests a Dual Role of an Odorant-Binding Protein in Two Sibling Helicoverpa Species. PLoS ONE 2012, 7, e30040.
- McKenzie, S.K.; Oxley, P.R.; Kronauer, D.J.C. Comparative genomics and transcriptomics in ants provide new insights into the evolution and function of odorant binding and chemosensory proteins. BMC Genom. 2014, 15, 1–14.
- Manoharan, M.; Chong, M.N.F.; Vaïtinadapoulé, A.; Frumence, E.; Sowdhamini, R.; Offmann, B. Comparative Genomics of Odorant Binding Proteins in Anopheles gambiae, Aedes aegypti, and Culex quinquefasciatus. Genome Biol. Evol. 2013, 5, 163–180.
- Sánchez-Gracia, A.; Vieira, F.G.; Rozas, J. Molecular evolution of the major chemosensory gene families in insects. Heredity 2009, 103, 208–216.
- Sun, J.S.; Xiao, S.; Carlson, J.R. The diverse small proteins called odorant-binding proteins. R. Soc. Open Biol. 2018, 8, 180208.
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 2017, 93, 184–200.
- Vieira, F.G.; Rozas, J. Comparative Genomics of the Odorant-Binding and Chemosensory Protein Gene Families across the Arthropoda: Origin and Evolutionary History of the Chemosensory System. Genome Biol. Evol. 2011, 3, 476–490.
- Eyun, S.-I.; Soh, H.Y.; Posavi, M.; Munro, J.B.; Hughes, D.S.; Murali, S.C.; Qu, J.; Dugan, S.; Lee, S.L.; Chao, H.; et al. Evolutionary History of Chemosensory-Related Gene Families across the Arthropoda. Mol. Cell Proteom. 2017, 34, 1838–1862.
- Epelosi, P.; Eiovinella, I.; Efelicioli, A.; Dani, F.R. Soluble proteins of chemical communication: An overview across arthropods. Front. Physiol. 2014, 5, 320.
- Steinbrecht, R.A.; Ozaki, M.; Ziegelberger, G. Immunocytochemical localization of pheromone-binding protein in moth antennae. Cell Tissue Res. 1992, 270, 287–302.
- Larter, N.K.; Sun, J.S.; Carlson, J.R. Organization and function of Drosophila odorant binding proteins. eLife 2016, 5.
- Yasukawa, J.; Tomioka, S.; Aigaki, T.; Matsuo, T. Evolution of expression patterns of two odorant-binding protein genes, Obp57d and Obp57e, in Drosophila. Gene 2010, 467, 25–34.
- Jeong, Y.T.; Shim, J.; Oh, S.R.; Yoon, H.I.; Kim, C.H.; Moon, S.J.; Montell, C. An Odorant-Binding Protein Required for Suppression of Sweet Taste by Bitter Chemicals. Neuron 2013, 79, 725–737.
- Rihani, K.; Fraichard, S.; Chauvel, I.; Poirier, N.; Delompré, T.; Neiers, F.; Tanimura, T.; Ferveur, J.-F.; Briand, L. A conserved odorant binding protein is required for essential amino acid detection in Drosophila. Commun. Biol. 2019, 2, 1–10.
- Pikielny, C.; Hasan, G.; Rouyer, F.; Rosbash, M. Members of a family of drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron 1994, 12, 35–49.
- Shanbhag, S.; Hekmat-Scafe, D.; Kim, M.-S.; Park, S.-K.; Carlson, J.; Pikielny, C.; Smith, D.; Steinbrecht, R. Expression mosaic of odorant-binding proteins inDrosophila olfactory organs. Microsc. Res. Tech. 2001, 55, 297–306.
- Zhu, J.; Iovinella, I.; Dani, F.R.; Liu, Y.-L.; Huang, L.-Q.; Liu, Y.; Wang, C.-Z.; Pelosi, P.; Wang, G. Conserved chemosensory proteins in the proboscis and eyes of Lepidoptera. Int. J. Biol. Sci. 2016, 12, 1394–1404.
- Chen, X.-F.; Xu, L.; Zhang, Y.-X.; Wei, D.; Wang, J.-J.; Jiang, H.-B. Genome-wide identification and expression profiling of odorant-binding proteins in the oriental fruit fly, Bactrocera dorsalis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 31, 100605.
- Pelletier, J.; Leal, W.S. Genome Analysis and Expression Patterns of Odorant-Binding Proteins from the Southern House Mosquito Culex pipiens quinquefasciatus. PLoS ONE 2009, 4, e6237.
- Sparks, J.T.; Bohbot, J.D.; Dickens, J.C. The genetics of chemoreception in the labella and tarsi of Aedes aegypti. Insect Biochem. Mol. Biol. 2014, 48, 8–16.
- Dippel, S.; Oberhofer, G.; Kahnt, J.; Gerischer, L.; Opitz, L.; Schachtner, J.; Stanke, M.; Schütz, S.; Wimmer, E.A.; Angeli, S. Tissue-specific transcriptomics, chromosomal localization, and phylogeny of chemosensory and odorant binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more general functions. BMC Genom. 2014, 15, 1–14.
- Pregitzer, P.; Zielonka, M.; Eichhorn, A.-S.; Jiang, X.; Krieger, J.; Breer, H. Expression of odorant-binding proteins in mouthpart palps of the desert locustSchistocerca gregaria. Insect Mol. Biol. 2019, 28, 264–276.
- Koganezawa, M.; Shimada, I. Novel odorant-binding proteins expressed in the taste tissue of the fly. Chem. Senses 2002, 27, 319–332.
- Galindo, K.; Smith, D.P. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics 2001, 159, 1059–1072.
- Li, Z.-X.; Pickett, J.A.; Field, L.M.; Zhou, J.-J. Identification and expression of odorant-binding proteins of the malaria-carrying mosquitoesAnopheles gambiae andAnopheles arabiensis. Arch. Insect Biochem. Physiol. 2005, 58, 175–189.
- Mitaka, H.; Matsuo, T.; Miura, N.; Ishikawa, Y. Identification of odorant-binding protein genes from antennal expressed sequence tags of the onion fly, Delia antiqua. Mol. Biol. Rep. 2010, 38, 1787–1792.
- Zheng, W.; Peng, W.; Zhu, C.; Zhang, Q.; Saccone, G.; Zhang, H. Identification and Expression Profile Analysis of Odorant Binding Proteins in the Oriental Fruit Fly Bactrocera dorsalis. Int. J. Mol. Sci. 2013, 14, 14936–14949.
- Gu, T.; Huang, K.; Tian, S.; Sun, Y.; Li, H.; Chen, C.; Hao, D. Antennal transcriptome analysis and expression profiles of odorant binding proteins in Clostera restitura. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 29, 211–220.
- Gu, S.-H.; Wang, S.-P.; Zhang, X.-Y.; Wu, K.-M.; Guo, Y.-Y.; Zhou, J.-J.; Zhang, Y.-J. Identification and tissue distribution of odorant binding protein genes in the lucerne plant bug Adelphocoris lineolatus (Goeze). Insect Biochem. Mol. Biol. 2011, 41, 254–263.
- Calvello, M.; Brandazza, A.; Navarrini, A.; Dani, F.; Turillazzi, S.; Felicioli, A.; Pelosi, P. Expression of odorant-binding proteins and chemosensory proteins in some Hymenoptera. Insect Biochem. Mol. Biol. 2005, 35, 297–307.
- Chintapalli, V.R.; Wang, J.; Dow, J.A.T. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 2007, 39, 715–720.
- Takemori, N.; Yamamoto, M.-T. Proteome mapping of the Drosophila melanogaster male reproductive system. Proteomics 2009, 9, 2484–2493.
- Prokupek, A.M.; Eyun, S.-I.; Ko, L.; Moriyama, E.N.; Harshman, L.G. Molecular evolutionary analysis of seminal receptacle sperm storage organ genes of Drosophila melanogaster. J. Evol. Biol. 2010, 23, 1386–1398.
- Sun, J.S.; Larter, N.K.; Chahda, J.S.; Rioux, D.; Gumaste, A.; Carlson, J.R. Humidity response depends on the small soluble protein Obp59a in Drosophila. eLife 2018, 7.
- Park, S.-K.; Shanbhag, S.; Dubin, A.; De Bruyne, M.; Wang, Q.; Yu, P.; Shimoni, N.; D’Mello, S.; Carlson, J.; Harris, G.; et al. Inactivation of olfactory sensilla of a single morphological type differentially affects the response ofDrosophila to odors. J. Neurobiol. 2002, 51, 248–260.
- Buchon, N.; Osman, D.; David, F.P.; Fang, H.Y.; Boquete, J.-P.; Deplancke, B.; Lemaitre, B. Morphological and Molecular Characterization of Adult Midgut Compartmentalization in Drosophila. Cell Rep. 2013, 3, 1725–1738.
- Iovinella, I.; Dani, F.R.; Niccolini, A.; Sagona, S.; Michelucci, E.; Gazzano, A.; Turillazzi, S.; Felicioli, A.; Pelosi, P. Differential Expression of Odorant-Binding Proteins in the Mandibular Glands of the Honey Bee According to Caste and Age. J. Proteome Res. 2011, 10, 3439–3449.
- Findlay, G.D.; Yi, X.; Maccoss, M.J.; Swanson, W.J. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 2008, 6, e178.
- Chapman, T. The Soup in My Fly: Evolution, Form and Function of Seminal Fluid Proteins. PLoS Biol. 2008, 6, e179.
- Sepil, I.; Hopkins, B.R.; Dean, R.; Thézénas, M.-L.; Charles, P.D.; Konietzny, R.; Fischer, R.; Kessler, B.M.; Wigby, S. Quantitative Proteomics Identification of Seminal Fluid Proteins in Male Drosophila melanogaster. Mol. Cell. Proteom. 2019, 18, S46–S58.
- Li, S.; Picimbon, J.-F.; Ji, S.; Kan, Y.; Chuanling, Q.; Zhou, J.-J.; Pelosi, P. Multiple functions of an odorant-binding protein in the mosquito Aedes aegypti. Biochem. Biophys. Res. Commun. 2008, 372, 464–468.
- Sirot, L.K.; Poulson, R.L.; McKenna, M.C.; Girnary, H.; Wolfner, M.F.; Harrington, L.C. Identity and transfer of male reproductive gland proteins of the dengue vector mosquito, Aedes aegypti: Potential tools for control of female feeding and reproduction. Insect Biochem. Mol. Biol. 2008, 38, 176–189.
- Baer, B.; Zareie, R.; Paynter, E.; Poland, V.; Millar, A.H. Seminal fluid proteins differ in abundance between genetic lineages of honeybees. J. Proteom. 2012, 75, 5646–5653.
- Xu, J.; Baulding, J.; Palli, S.R. Proteomics of Tribolium castaneum seminal fluid proteins: Identification of an angiotensin-converting enzyme as a key player in regulation of reproduction. J. Proteom. 2013, 78, 83–93.
- Costa-Da-Silva, A.L.; Kojin, B.B.; Marinotti, O.; James, A.A.; Capurro, M.L. Expression and accumulation of the two-domain odorant-binding protein AaegOBP45 in the ovaries of blood-fed Aedes aegypti. Parasites Vectors 2013, 6, 364.
- Marinotti, O.; Ngo, T.; Kojin, B.B.; Chou, S.-P.; Nguyen, B.; Juhn, J.; Carballar-Lejarazú, R.; Marinotti, P.N.; Jiang, X.; Walter, M.F.; et al. Integrated proteomic and transcriptomic analysis of the Aedes aegypti eggshell. BMC Dev. Biol. 2014, 14, 15.
- Amenya, D.A.; Chou, W.; Li, J.; Yan, G.; Gershon, P.D.; James, A.A.; Marinotti, O. Proteomics reveals novel components of the Anopheles gambiae eggshell. J. Insect Physiol. 2010, 56, 1414–1419.
- Andersson, M.N.; Videvall, E.; Walden, K.K.O.; Harris, M.O.; Robertson, H.M.; Löfstedt, C. Sex- and tissue-specific profiles of chemosensory gene expression in a herbivorous gall-inducing fly (Diptera: Cecidomyiidae). BMC Genom. 2014, 15, 501.
- Xu, W.; Cornel, A.J.; Leal, W.S. Odorant-Binding Proteins of the Malaria Mosquito Anopheles funestus sensu stricto. PLoS ONE 2010, 5, e15403.
- Heavner, M.E.; Gueguen, G.; Rajwani, R.; Pagan, P.E.; Small, C.; Govind, S. Partial venom gland transcriptome of a Drosophila parasitoid wasp, Leptopilina heterotoma, reveals novel and shared bioactive profiles with stinging Hymenoptera. Gene 2013, 526, 195–204.
- Wang, L.; Zhu, J.-Y.; Qian, C.; Fang, Q.; Ye, G.-Y. VENOM OF THE PARASITOID WASPPteromalus puparumCONTAINS AN ODORANT BINDING PROTEIN. Arch. Insect Biochem. Physiol. 2015, 88, 101–110.
- Li, R.; Zhang, L.; Fang, Y.; Han, B.; Lu, X.; Zhou, T.; Feng, M.; Li, J. Proteome and phosphoproteome analysis of honeybee (Apis mellifera) venom collected from electrical stimulation and manual extraction of the venom gland. BMC Genom. 2013, 14, 766.
- Ishida, Y.; Ishibashi, J.; Leal, W.S. Fatty Acid Solubilizer from the Oral Disk of the Blowfly. PLoS ONE 2013, 8, e51779.
- Ribeiro, S.P.; Genta, F.A.; Sorgine, M.H.F.; Logullo, R.; Mesquita, R.D.; Paiva-Silva, G.O.; Majerowicz, D.; Medeiros, M.; Koerich, L.; Terra, W.R.; et al. An Insight into the Transcriptome of the Digestive Tract of the Bloodsucking Bug, Rhodnius prolixus. PLoS Negl. Trop. Dis. 2014, 8, e2594.
- Smartt, C.T.; Erickson, J.S. Bloodmeal-induced differential gene expression in the disease vector culex nigripalpus (Diptera: Culicidae). J. Med. Entomol. 2008, 45, 326–330.
- Smartt, C.T.; Erickson, J.S. Expression of a Novel Member of the Odorant-Binding Protein Gene Family in Culex nigripalpus (Diptera: Culicidae). J. Med. Entomol. 2009, 46, 1376–1381.
- Scully, E.D.; Geib, S.M.; Mason, C.J.; Carlson, J.E.; Tien, M.; Chen, H.-Y.; Harding, S.; Tsai, C.-J.; Hoover, K. Host-plant induced changes in microbial community structure and midgut gene expression in an invasive polyphage (Anoplophora glabripennis). Sci. Rep. 2018, 8, 1–16.
- Benoit, J.B.; Vigneron, A.; Broderick, N.A.; Wu, Y.; Sun, J.S.; Carlson, J.R.; Aksoy, S.; Weiss, B.L. Symbiont-induced odorant binding proteins mediate insect host hematopoiesis. eLife 2017, 6.
- Gonzalez, D.; Rihani, K.; Neiers, F.; Poirier, N.; Fraichard, S.; Gotthard, G.; Chertemps, T.; Maïbèche, M.; Ferveur, J.-F.; Briand, L. The Drosophila odorant-binding protein 28a is involved in the detection of the floral odour ß-ionone. Cell. Mol. Life Sci. 2019, 77, 2565–2577.
