The coronavirus disease 2019 (COVID-19) outbreak caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was proclaimed a global pandemic in March 2020. Reducing the dissemination rate, in particular by tracking the infected people and their contacts, is the main instrument against infection spreading. Therefore, the creation and implementation of fast, reliable and responsive methods suitable for the diagnosis of COVID-19 are required. These needs can be fulfilled using affinity sensors, which differ in applied detection methods and markers that are generating analytical signals. Recently, nucleic acid hybridization, antigen-antibody interaction, and change of reactive oxygen species (ROS) level are mostly used for the generation of analytical signals, which can be accurately measured by electrochemical, optical, surface plasmon resonance, field-effect transistors, and some other methods and transducers.
- COVID-19
- SARS-CoV-2 virus
- RNA analysis
- bioelectrochemistry
- biosensors
- electrochemical immunosensors
- antigen-antibody interaction
- immune complex
- molecularly imprinted polymers (MIPs)
- surface modification by immobilization of biomolecules
1. Introduction
The spreading of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is causing coronavirus disease 2019 (COVID-19) was declared as a global pandemic in March 2020. The main threat of the pandemic is the overloading of the health systems. The key tool against infection spreading is decreasing its distribution rate, in particular by monitoring the infected people and their contacts. For the successful control, the primary step is the detection of SARS-CoV-2 in an organism. Hence, the development and introduction of rapid, precise, and sensitive detection methods are required. For a better understanding of the existing detection method principles, it is worth dwelling in more detail on the structure of SARS-CoV-2, its life cycle and the induced host response.
2. Affinity Biosensors for COVID-19 Diagnosis
SARS-CoV-2 infection can be identified using affinity biosensors [1]. Several different types of signal transduction systems can be applied, which include electrochemical, optical, piezoelectric and some others. Electrochemical affinity biosensors are the most prevalent in biomedical applications due to their cheapness, ease and facility of mass manufacture [1].
6.1. Affinity Biosensors for the Determination of SARS-CoV-2 RNA
An electrochemical DNA/RNA biosensor employs the hybridization of single-stranded nucleic acid (NA) with the complementary strand as a source of the electrochemical signal [2]. The affinity biosensor includes a biorecognition element consisting of the capture NA specifically interacting with the target NA, and the signal transducer where the identification event is transformed into an electrical signal [3] (Figure 1). The detection of specific hybridization of two complementary strands of NA is the key of the affinity biosensor working principle [4][5]. Sometimes additional reporter probes, which are marked with signaling compounds, are used. The reaction of hybridization occurs on an electrode or in a solution [6].
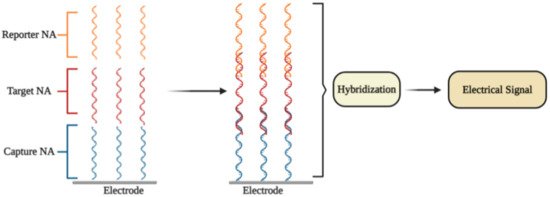
Figure 1. The general principle of electrochemical affinity biosensors for the detection of specific nucleic acid sequences.
In some electrochemical sensors, NA hybridization [2] includes an electrochemical reaction, which is further used for the quantitation of the detected NA fragment concentration and thus to the concentration of SARS-CoV-2 virus. Electrochemical NA biosensors are classified according to the types of reporter NA (label-free or labeled) and through the signal generation principle (reagent-free or reagent-dependent) [7].
The selective identification of a low amount of DNA and/or RNA copies in specimens is the most important task for electrochemical NA biosensors. The choice of the most efficient signal amplification method is the key aspect that is used to resolve this task. The molecular approaches are classified into (1) NA-based amplification methods (enzyme-mediated isothermal amplification of NA), (2) nanomaterials-based methods (large surface area for the loading of capture NA; nanomaterials as reporter probes), and (3) enzyme-mediated signal amplification (enzymes are connected with NA hybridization system) [7]. Different electrochemical methods are employed for the quantitation of amplified signals, namely, electrochemical impedance spectroscopy (EIS) [8][9], chronoamperometry [10], pulsed amperometric detection [2][11], square wave voltammetry [12], differential pulse voltammetry (DPV) [13], and cyclic voltammetry (CV) [14][15].
6.1.1. DPV-Based Affinity Biosensors
Some researchers presented ultrasensitive DPV-based detection technology using calixarene functionalized graphene oxide for targeting RNA of SARS-CoV-2 [16]. It was affirmed that the technology identifies RNA of SARS-CoV-2 avoiding amplification and reverse transcription stages by employing a portable electrochemical smartphone. The biosensor consists of a capture probe, target sequence, label probe, and an auxiliary probe [17]. The capture probe is complementary to the 5′-terminal of the target sequence, while the label probe is complementary to the 3′-terminal; two different label probe areas have complementary sequences to 5′- and 3′-regions of the auxiliary probe [17][18]. Commonly, each label probe was marked with only one signal compound that led to a low current signal. Hence, it is assumed that transferring the label probe of signaling molecules to other materials or compounds may help to increase the sensitivity [16]. The LOD in the clinical sample is 200 copies/mL, from which it follows that only two copies (10 μL) of all viral RNA copies are needed per analysis. The sensitivity for samples from confirmed COVID-19 patients was 85.5%. [16].
It is worth noting that there are some investigations concerning the potential use of G-quadruplex-based biosensors in COVID-19 diagnosis [19]. G-quadruplex (GQ) is a guanine-rich DNA/RNA sequence, which is folded into four-stranded secondary structures. These structures take part in crucial genome functions such as transcription, replication, and genome stability [20]. Recently, 25 putative G-quadruplex-forming sequences (PQSs) in the genome of SARS-CoV-2 virus were recognized [21]. The PQSs are situated in the ORF1ab, ORF3a, S-, M-, and N-genes of SARS-CoV-2 [19]. Some of the found PQSs are observed in a wide range of coronaviruses, while the main two PQSs, which generate RNA G-quadruplex structures, are strictly observed only in a limited range of viruses. Moreover, a straight interaction between G-quadruplex of coronavirus and viral helicase (nsp13) was obtained by microscale thermophoresis. The results of molecular docking-based modeling suggest that nsp13 alters the G-quadruplex structure. The helicase allows the guanine bases to go out of the guanine quartet planes, therefore, simplifying their unfolding [21]. Thus, RNA G-quadruplex sequences of SARS-CoV-2 could be used for the design of affinity-sensors, which are based on the identification of the viral helicase protein, nsp13 [21].
Fluorescence quenching is a powerful technique for the design of affinity biosensors [22]. One type of biosensor for the determination of enzymes based on fluorescence quenching by G-quadruplex has been reported recently [23]. Guanine at a lower oxidation state can act as the electron donor, while the fluorescence able group acts as an acceptor, which further produces a signal [24].
6.1.2. Plasmonics-Based Affinity Biosensors
The primary concept of plasmonic biosensors is based on the distribution of surface plasmons lengthwise the interface of the thin metallic layer (usually noble metals), and dielectric [25]. This method consists of real-time monitoring changes of the refractive index of the medium surrounding the sensor surface during the interactions between the target biocompound and the immobilized biorecognition element [26][27][28][29]. Most plasmonic biosensors are built on the basics of surface plasmon resonance (SPR) [25][30]. Interactions occur on the surface that is suitable for observation SPR-based signals in two different modes: (1) bulk SPR signal and (2) localized SPR (LSPR) signal. Both effects rely on the refractive index of the ambient media to evoke spectral shifts. Nevertheless, the distinction between SPR and LSPR is defined by the dimensions of applied plasmonic nanomaterials [31].
It was reported that a dual-functional plasmonic biosensor incorporating the plasmonic photothermal (PPT) effect and LSPR sensing transmission enables the development of an alternative approach for SARS-CoV-2 virus detection, where the detection is provided through the hybridization of complementary NA with one NA immobilized on the surface of the gold nanoislands (AuNIs). The LSPR and PPT effects were utilized mutually to increase the signal. The LOD of this assay for the RdRp gene was 0.22 pM. The specificity, the discrimination between the RdRp gene of SARS-CoV and SARS-CoV-2, can be precisely established by onsite PPT improvement on gold AuNIs-based chips [32].
Plasmonic biosensing has technological benefits including the possibility of a combination of SPR with electrochemical, and electroassisted chemiluminescence methods [33]. Moreover, some nanomaterials were applied to establish the optical aperture and reach very sensitive virus identification by SPR method combined with colorimetric and fluorescence determination based approaches [25]. Kinds of plasmonic nanomaterials can alter from metallic nanoparticles and quantum dots to graphene nanostructures [34][35][36][37].
6.2. Immunosensors for Determination of SARS-CoV-2 Proteins
6.2.1. Field-Effect Transistor Based Immunosensors
It was reported that a Field-effect transistor (FET)-based biosensor enables the real-time detection of SARS-CoV-2 in clinical specimens. The device was manufactured by covering the graphene plates of the FET with an antibody produced as a response to the SARS-CoV-2 S-protein. The antibody was fixed on the surface of the biosensor by 1-pyrenebutyric acid N-hydroxysuccinimide ester (PBASE) (Figure 2). The cultured virus, antigen protein, and nasopharyngeal swab samples from an infected person have been utilized for the assessment of the efficiency of the immunosensor. It was determined that the FET immunosensor enables the detection of the S-protein at a concentration of 1 fg/mL in PBS and 100 fg/mL in the transport medium, whereas LOD for SARS-CoV-2 was 1.6 × 101 pfu/mL in culture medium and 2.42 × 102 copies/mL in clinical specimens [38].
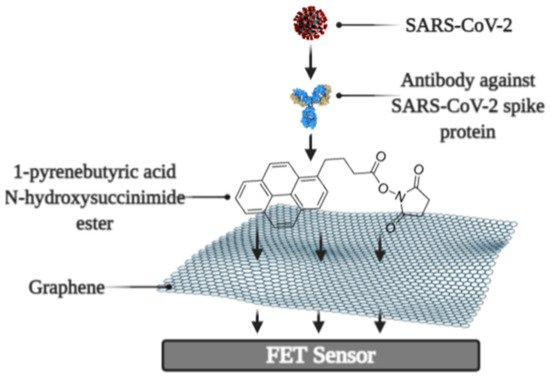
Figure 2. Schematic representation of field-effect transistor-based immunosensor for SARS-CoV-2 detection.
6.2.2. Quartz Crystal Microbalance Based Biosensors
The quartz crystal microbalance (QCM) can be successfully applied for the development of affinity biosensors [39]. In QCM-based approach, the binding with the viral S-protein occurs on the engineered quartz crystal surface covered by self-assembled monolayer (SAM), and the detection is carried out by QCM. A very simple approach for the determination of proteins is to exploit rather basic surface properties such as hydrophobicity, which is one of the key properties of the working surface of such an analytical system since the increasing the wettability of the surface leads to the increased surface concentration of proteins [40][41]. For this purpose, SAMs with a varied range of hydrophobicity, which is controlled by surface functional groups, were investigated and developed [42]. The SAMs, which have terminal –COOH and –CH3 groups, have been shown as the most suitable for the specific and strong binding of SARS-CoV-2 S-protein [43]. The main working principle of the QCM is altering (decreasing) the frequency of the vibrating quartz crystal with the increasing the adsorbed mass [44]. Therefore, QCM-based techniques enable performing sensitive, rapid, and label-free tests [44][45].
One more type of affinity biosensor, which is very promising for the determination of virus-induced diseases, is the ultrasound transducer-based immunosensors, e.g.: capacitive micromachined ultrasound transducer (cMUT) was applied in immunosensors for the detection of specific antibodies against some virus proteins [46]. Moreover, the ultrasound-based test allows performing the SARS-CoV-2 virus detection in the gas phase (ultrasonator-produced viral aerosol) [47], while the vast majority of the assays are designed for the solution.
6.2.3. Molecularly Imprinted Polymer Based Electrochemical Affinity Sensors
In affinity sensors, the target protein is detected on the surface of the device, thus the design of the surface with appropriate protein recognition properties is required for the development of such sensors. For this purpose, molecularly imprinted polymers (MIPs) can be very efficiently applied [9][48][49][50][51]. The advantage of molecularly imprinted sensors is that they are cheaper and more stable, and can be based on protein-imprinted polymers such as polypyrrole [52] and some other electrochemically deposited polymers [53][54][55]. Various signal determination methods can be applied in the design of MIP-based sensors, but mostly potentiodynamic electrochemical techniques [52] or QCM-based [39][56] approaches are used for this purpose.
Development and application of MIPs in sensor design is reasonable because MIPs can be developed for small and low molecular weight molecules [14][57]. The efficiency of MIPs for the determination of some virus proteins was also demonstrated [52] and this technology recently was applied for the development of a molecularly imprinted poly-m-phenylenediamine based electrochemical sensors for the detection of SARS-CoV-2 proteins, namely, N-protein [58]. The sensor represented a disposable MIP-modified thin film electrode possessing selectivity to N-protein. Electrochemical signal was observed by DPV and a linear response to N-protein was up to 111 fM with a detection and quantification limit of 15 fM and 50 fM [58].
It should be noted that even some short DNA-based oligomers can be determined by MIP-based sensors [49], which makes MIP-based sensors attractive for DNA and probably for RNA fragment determination. Due to the rather low price of MIPs in comparison to that of antibodies, MIP-related research area is of particular interest and, therefore, MIPs potentially can replace antibodies during the design of various bioanalytical systems and immunosensors.
6.3. Ellipsometry and SPR Based Immunosensors
Optical ellipsometry-based techniques have great potential to be applied in the design of various immunosensors [59]. Comparing with other existing methods (ELISA, RT-PCR, indirect fluorescent, western blot) of SARS-CoV detection, the imaging ellipsometry-based approach has established itself as a direct, nondestructive, quick, label-free, simple, and low-cost technique [60].
Recently, spectroscopic ellipsometry (SE) in total internal reflection mode (TIRE) was applied for the monitoring the kinetic of interactions between on SAM-modified gold disk immobilized SARS-CoV-2 N-protein and antibodies against it [61]. TIRE allowed detecting biomolecules mass changes at solid-liquid interface by phase shift measurement. The high sensitivity of SE TIRE was attained with the support of SPR, what enabled the registration of two kinetic curves Ψ(t) and Δ(t) simultaneously [62][63]. It was reported, that antigen-antibody complex is strongly bound and the complex formation has very strict orientation requirements, which was established by meaning of mathematical model building [61]. The main working element of the sensor is the piezoelectric resonator, on which an antigen or antibody is immobilized using SAM-based technology. Incidentally, the application of antibody fragments seems to be a very promising approach for the development of sensors for the determination of virus proteins because it enables increasing the surface concertation of sites that are selective to virus proteins [64][65]
There are some other researches dedicated to the development of biosensors with the potential application for the determination of SARS-CoV-2 infection, based on the antigen-antibody interactions. In order to exploit such interactions, an electrochemical biosensor based on electrode covered with a SAM and specific-antibodies against SARS-CoV-2 proteins, was designed [66].
6.4. Photoluminescence-Based Immunosensors
Photoluminescence is a very sensitive technique that can be applied in the design of various affinity biosensors for the determination of pathological cells [67] and virus-induced diseases [68][69][70]. Some researchers designed a split luciferase (spLUC) based antibody test that is showing itself as simple (not need ‘washing’, two-stage of reagent addition, rapid (less than 5 min), reliable (≥98%), low-volume specimen (1 µL for 1 reaction), inexpensive and solution-based quantitative approach to identify antibodies against S- and N- proteins of SARS-CoV-2 [71]. The biosensor was designed by merging small BiT (SmBiT) and large BiT (LgBiT) fragments [72] of Nanoluciferase (NanoLuc) to viral protein antigens. The immunoglobulin has two antigen-binding sites, thus the outcome of incubating 1:1 mixture of SmBiT and LgBiT with serum will be the coupling of one antigen-binding site with LgBiT and another site with SmBiT. The fixing of LgBiT and SmBiT fragments leads to the reduction of NanoLuc enzyme for the following luminescence-based identification (Figure 3).
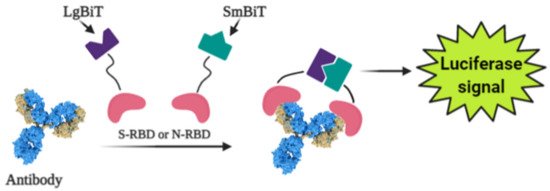
Figure 3. Scheme of the general working principle of split luciferase based immunosensor.
Sensors based on S- and N-proteins of the SARS-CoV-2 were designed because SARS-CoV-2 infected patients contain antibodies, which are primarily addressed to S- and N-protein epitopes [73][74]. The sensor-based on genetically engineered S-protein containing merged RBD with NanoLuc fragments, whereas for the creation of N-protein-based sensor N-terminal sequence was utilized. The ordinary differential equation modeling was executed to describe the ratio between signal intensity and immunoglobulin concentration and it was shown that there was a linear correlation between the specific antibody concentration and luciferase signal. The sensor showed sensitivity of 89% towards S-protein and 98% towards N-protein [71].
This entry is adapted from the peer-reviewed paper 10.3390/mi12040390
References
- Cui, F.; Zhou, H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020, 165, 112349.
- Ramanaviciene, A.; Ramanavicius, A. Pulsed amperometric detection of DNA with an ssDNA/polypyrrole-modified electrode. Anal. Bioanal. Chem. 2004, 379, 287–293.
- Drummond, T.G.; Hill, M.G.; Barton, J.K. Electrochemical DNA sensors. Nat. Biotechnol. 2003, 21, 1192–1199.
- Zhang, D.Y.; Chen, S.X.; Yin, P. Optimizing the specificity of nucleic acid hybridization. Nat. Chem. 2012, 4, 208–214.
- Pellitero, M.A.; Shaver, A.; Arroyo-Currás, N. Critical Review—Approaches for the Electrochemical Interrogation of DNA-Based Sensors: A Critical Review. J. Electrochem. Soc. 2020, 167, 037529.
- Trotter, M.; Borst, N.; Thewes, R.; von Stetten, F. Review: Electrochemical DNA sensing—Principles, commercial systems, and applications. Biosens. Bioelectron. 2020, 154, 112069.
- Santhanam, M.; Algov, I.; Alfonta, L. DNA/RNA Electrochemical Biosensing Devices a Future Replacement of PCR Methods for a Fast Epidemic Containment. Sensors 2020, 20, 4648.
- Ramanavicius, A.; Finkelsteinas, A.; Cesiulis, H.; Ramanaviciene, A. Electrochemical impedance spectroscopy of polypyrrole based electrochemical immunosensor. Bioelectrochemistry 2010, 79, 11–16.
- Ratautaite, V.; Janssens, S.D.; Haenen, K.; Nesládek, M.; Ramanaviciene, A.; Baleviciute, I.; Ramanavicius, A. Molecularly Imprinted Polypyrrole Based Impedimentric Sensor for Theophylline Determination. Electrochim. Acta 2014, 130, 361–367.
- German, N.; Ramanavicius, A.; Ramanaviciene, A. Electrochemical deposition of gold nanoparticles on graphite rod for glucose biosensing. Sens. Actuators B Chem. 2014, 203, 25–34.
- Ramanavicius, A.; Oztekin, Y.; Ramanaviciene, A. Electrochemical formation of polypyrrole-based layer for immunosensor design. Sens. Actuators B Chem. 2014, 197, 237–243.
- Oztekin, Y.; Yazicigil, Z.; Ramanaviciene, A.; Ramanavicius, A. Square wave voltammetry based on determination of copper (II) ions by polyluteolin- and polykaempferol-modified electrodes. Talanta 2011, 85, 1020–1027.
- Deshmukh, M.A.; Patil, H.K.; Bodkhe, G.A.; Yasuzawa, M.; Koinkar, P.; Ramanaviciene, A.; Shirsat, M.D.; Ramanavicius, A. EDTA-modified PANI/SWNTs nanocomposite for differential pulse voltammetry based determination of Cu(II) ions. Sens. Actuators B Chem. 2018, 260, 331–338.
- Ramanavicius, S.; Ramanavicius, A. Conducting Polymers in the Design of Biosensors and Biofuel Cells. Polymers 2020, 13, 49.
- Samukaite-Bubniene, U.; Valiuniene, A.; Bucinskas, V.; Genys, P.; Ratautaite, V.; Ramanaviciene, A.; Aksun, E.; Tereshchenko, A.; Zeybek, B.; Ramanavicius, A. Towards supercapacitors: Cyclic voltammetry and fast Fourier transform electrochemical impedance spectroscopy based evaluation of polypyrrole electrochemically deposited on the pencil graphite electrode. Colloid Surf. A Physicochem. Eng. Asp. 2021, 610.
- Zhao, H.; Liu, F.; Xie, W.; Zhou, T.C.; OuYang, J.; Jin, L.; Li, H.; Zhao, C.Y.; Zhang, L.; Wei, J.; et al. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sens. Actuators B Chem. 2021, 327, 128899.
- Chen, X.; Lin, Y.H.; Li, J.; Lin, L.S.; Chen, G.N.; Yang, H.H. A simple and ultrasensitive electrochemical DNA biosensor based on DNA concatamers. Chem. Commun. 2011, 47, 12116–12118.
- Wang, J.; Shi, A.; Fang, X.; Han, X.; Zhang, Y. An ultrasensitive supersandwich electrochemical DNA biosensor based on gold nanoparticles decorated reduced graphene oxide. Anal. Biochem. 2015, 469, 71–75.
- Xi, H.; Juhas, M.; Zhang, Y. G-quadruplex based biosensor: A potential tool for SARS-CoV-2 detection. Biosens. Bioelectron. 2020, 167, 112494.
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136.
- Ji, D.; Juhas, M.; Tsang, C.M.; Kwok, C.K.; Li, Y.; Zhang, Y. Discovery of G-quadruplex-forming sequences in SARS-CoV-2. Brief. Bioinf. 2020.
- Ramanavicius, A.; Kurilcik, N.; Jursenas, S.; Finkelsteinas, A.; Ramanaviciene, A. Conducting polymer based fluorescence quenching as a new approach to increase the selectivity of immunosensors. Biosens. Bioelectron. 2007, 23, 499–505.
- Wu, K.; Ma, C.; Deng, Z.; Fang, N.; Tang, Z.; Zhu, X.; Wang, K. Label-free and nicking enzyme-assisted fluorescence signal amplification for RNase H determination based on a G-quadruplexe/thioflavin T complex. Talanta 2018, 182, 142–147.
- Ying, L.; Green, J.J.; Li, H.; Klenerman, D.; Balasubramanian, S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 2003, 100, 14629–14634.
- Mauriz, E. Recent Progress in Plasmonic Biosensing Schemes for Virus Detection. Sensors 2020, 20, 4745.
- Homola, J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 2008, 108, 462–493.
- Brolo, A.G. Plasmonics for future biosensors. Nat. Photonics 2012, 6, 709–713.
- Sipova, H.; Homola, J. Surface plasmon resonance sensing of nucleic acids: A review. Anal. Chim. Acta 2013, 773, 9–23.
- Li, Z.; Leustean, L.; Inci, F.; Zheng, M.; Demirci, U.; Wang, S. Plasmonic-based platforms for diagnosis of infectious diseases at the point-of-care. Biotechnol. Adv. 2019, 37, 107440.
- Kausaite-Minkstimiene, A.; Ramanavicius, A.; Ruksnaite, J.; Ramanaviciene, A. A surface plasmon resonance immunosensor for human growth hormone based on fragmented antibodies. Anal. Methods 2013, 5, 4757–4763.
- Li, M.; Cushing, S.K.; Wu, N. Plasmon-enhanced optical sensors: A review. Analyst 2015, 140, 386–406.
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277.
- Ramanaviciene, A.; German, N.; Kausaite-Minkstimiene, A.; Voronovic, J.; Kirlyte, J.; Ramanavicius, A. Comparative study of surface plasmon resonance, electrochemical and electroassisted chemiluminescence methods based immunosensor for the determination of antibodies against human growth hormone. Biosens. Bioelectron. 2012, 36, 48–55.
- Tran, V.T.; Zhou, H.; Kim, S.; Lee, J.; Kim, J.; Zou, F.; Kim, J.; Park, J.Y. Self-assembled magnetoplasmonic nanochain for DNA sensing. Sens. Actuators B Chem. 2014, 203, 817–823.
- Adegoke, O.; Park, E.Y. Gold Nanoparticle-Quantum Dot Fluorescent Nanohybrid: Application for Localized Surface Plasmon Resonance-induced Molecular Beacon Ultrasensitive DNA Detection. Nanoscale Res. Lett. 2016, 11, 523.
- Lee, J.; Takemura, K.; Park, E.Y. Plasmonic Nanomaterial-Based Optical Biosensing Platforms for Virus Detection. Sensors 2017, 17.
- Farzin, L.; Shamsipur, M.; Samandari, L.; Sheibani, S. HIV biosensors for early diagnosis of infection: The intertwine of nanotechnology with sensing strategies. Talanta 2020, 206, 120201.
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.H.; Choi, M.; Ku, K.B.; Lee, C.S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142.
- Plausinaitis, D.; Sinkevicius, L.; Samukaite-Bubniene, U.; Ratautaite, V.; Ramanavicius, A. Evaluation of electrochemical quartz crystal microbalance based sensor modified by uric acid-imprinted polypyrrole. Talanta 2020, 220, 121414.
- Disley, D.M.; Cullen, D.C.; You, H.-X.; Lowe, C.R. Covalent coupling of immunoglobulin G to self-assembled monolayers as a method for immobilizing the interfacial-recognition layer of a surface plasmon resonance immunosensor. Biosens. Bioelectron. 1998, 13, 1213–1225.
- Hasan, A.; Pattanayek, S.K.; Pandey, L.M. Effect of Functional Groups of Self-Assembled Monolayers on Protein Adsorption and Initial Cell Adhesion. ACS Biomater. Sci. Eng. 2018, 4, 3224–3233.
- Pandey, L.M.; Pattanayek, S.K. Relation between the Wetting Effect and the Adsorbed Amount of Water-Soluble Polymers or Proteins at Various Interfaces. J. Chem. Eng. Data 2013, 58, 3440–3446.
- Pandey, L.M. Design of engineered surfaces for prospective detection of SARS-CoV-2 using quartz crystal microbalance-based techniques. Expert Rev. Proteomics 2020, 17, 425–432.
- Pandey, L.M.; Pattanayek, S.K. Hybrid surface from self-assembled layer and its effect on protein adsorption. Appl. Surface Sci. 2011, 257, 4731–4737.
- Deng, T.; Li, J.S.; Huan, S.Y.; Yang, H.F.; Wang, H.; Shen, G.L.; Yu, R.Q. Quartz crystal microbalance bioaffinity sensor for biotin based on mixed self-assembled monolayers and metastable molecular complex receptor. Biosens. Bioelectron. 2006, 21, 1545–1552.
- Ramanaviciene, A.; Virzonis, D.; Vanagas, G.; Ramanavicius, A. Capacitive micromachined ultrasound transducer (cMUT) for immunosensor design. Analyst 2010, 135, 1531–1534.
- Zuo, B.; Li, S.; Guo, Z.; Zhang, J.; Chen, C. Piezoelectric immunosensor for SARS-associated coronavirus in sputum. Anal. Chem. 2004, 76, 3536–3540.
- Ramanaviciene, A.; Ramanavicius, A.; Finkelsteinas, A. Basic Electrochemistry Meets Nanotechnology: Electrochemical Preparation of Artificial Receptors Based on Nanostructured Conducting Polymer, Polypyrrole. J. Chem. Educ. 2006, 83, 1212–1214.
- Ratautaite, V.; Topkaya, S.N.; Mikoliunaite, L.; Ozsoz, M.; Oztekin, Y.; Ramanaviciene, A.; Ramanavicius, A. Molecularly Imprinted Polypyrrole for DNA Determination. Electroanalysis 2013, 25, 1169–1177.
- Viter, R.; Kunene, K.; Genys, P.; Jevdokimovs, D.; Erts, D.; Sutka, A.; Bisetty, K.; Viksna, A.; Ramanaviciene, A.; Ramanavicius, A. Photoelectrochemical Bisphenol S Sensor Based on ZnO-Nanoroads Modified by Molecularly Imprinted Polypyrrole. Macromol. Chem. Phys. 2019, 221.
- Syritski, V.; Reut, J.; Menaker, A.; Gyurcsányi, R.E.; Öpik, A. Electrosynthesized molecularly imprinted polypyrrole films for enantioselective recognition of l-aspartic acid. Electrochim. Acta 2008, 53, 2729–2736.
- Ramanaviciene, A.; Ramanavicius, A. Molecularly imprinted polypyrrole-based synthetic receptor for direct detection of bovine leukemia virus glycoproteins. Biosens. Bioelectron. 2004, 20, 1076–1082.
- Boroznjak, R.; Reut, J.; Tretjakov, A.; Lomaka, A.; Opik, A.; Syritski, V. A computational approach to study functional monomer-protein molecular interactions to optimize protein molecular imprinting. J. Mol. Recognit. 2017, 30.
- Menaker, A.; Syritski, V.; Reut, J.; Öpik, A.; Horváth, V.; Gyurcsányi, R.E. Electrosynthesized Surface-Imprinted Conducting Polymer Microrods for Selective Protein Recognition. Adv. Mater. 2009, 21, 2271–2275.
- Tretjakov, A.; Syritski, V.; Reut, J.; Boroznjak, R.; Volobujeva, O.; Öpik, A. Surface molecularly imprinted polydopamine films for recognition of immunoglobulin G. Microchim. Acta 2013, 180, 1433–1442.
- Syritski, V.; Reut, J.; Öpik, A.; Idla, K. Environmental QCM sensors coated with polypyrrole. Synth. Metals 1999, 102, 1326–1327.
- Ramanavicius, S.; Ramanavicius, A. Charge Transfer and Biocompatibility Aspects in Conducting Polymer-Based Enzymatic Biosensors and Biofuel Cells. Nanomaterials 2021, 11, 371.
- Raziq, A.; Kidakova, A.; Boroznjak, R.; Reut, J.; Opik, A.; Syritski, V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021, 178, 113029.
- Balevicius, Z.; Paulauskas, A.; Plikusiene, I.; Mikoliunaite, L.; Bechelany, M.; Popov, A.; Ramanavicius, A.; Ramanaviciene, A. Towards the application of Al2O3/ZnO nanolaminates in immunosensors: Total internal reflection spectroscopic ellipsometry based evaluation of BSA immobilization. J. Mater. Chem. C 2018, 6, 8778–8783.
- Qi, C.; Duan, J.Z.; Wang, Z.H.; Chen, Y.Y.; Zhang, P.H.; Zhan, L.; Yan, X.Y.; Cao, W.C.; Jin, G. Investigation of interaction between two neutralizing monoclonal antibodies and SARS virus using biosensor based on imaging ellipsometry. Biomed. Microdevices 2006, 8, 247–253.
- Plikusiene, I.; Maciulis, V.; Ramanaviciene, A.; Balevicius, Z.; Buzavaite-Verteliene, E.; Ciplys, E.; Slibinskas, R.; Simanavicius, M.; Zvirbliene, A.; Ramanavicius, A. Evaluation of Kinetics and Thermodynamics of Interaction between Immobilized SARS-CoV-2 Nucleoprotein and Specific Antibodies by Total Internal Reflection Ellipsometry. J. Colloid Interface Sci. 2021.
- Baleviciute, I.; Balevicius, Z.; Makaraviciute, A.; Ramanaviciene, A.; Ramanavicius, A. Study of antibody/antigen binding kinetics by total internal reflection ellipsometry. Biosens. Bioelectron. 2013, 39, 170–176.
- Arwin, H.; Poksinski, M.; Johansen, K. Total internal reflection ellipsometry: Principles and applications. Appl. Opt. 2004, 43, 3028–3036.
- Kausaite-Minkstimiene, A.; Ramanaviciene, A.; Kirlyte, J.; Ramanavicius, A. Comparative study of random and oriented antibody immobilization techniques on the binding capacity of immunosensor. Anal. Chem. 2010, 82, 6401–6408.
- Balevicius, Z.; Ramanaviciene, A.; Baleviciute, I.; Makaraviciute, A.; Mikoliunaite, L.; Ramanavicius, A. Evaluation of intact- and fragmented-antibody based immunosensors by total internal reflection ellipsometry. Sens. Actuators B Chem. 2011, 160, 555–562.
- Mayall, R.M.; Smith, C.A.; Hyla, A.S.; Lee, D.S.; Crudden, C.M.; Birss, V.I. Ultrasensitive and Label-Free Detection of the Measles Virus Using an N-Heterocyclic Carbene-Based Electrochemical Biosensor. ACS Sens. 2020, 5, 2747–2752.
- Tamashevski, A.; Harmaza, Y.; Viter, R.; Jevdokimovs, D.; Poplausks, R.; Slobozhanina, E.; Mikoliunaite, L.; Erts, D.; Ramanaviciene, A.; Ramanavicius, A. Zinc oxide nanorod based immunosensing platform for the determination of human leukemic cells. Talanta 2019, 200, 378–386.
- Tereshchenko, A.; Smyntyna, V.; Ramanavicius, A. Interaction mechanism between TiO2 nanostructures and bovine leukemia virus proteins in photoluminescence-based immunosensors. RSC Adv. 2018, 8, 37740–37748.
- Viter, R.; Savchuk, M.; Iatsunskyi, I.; Pietralik, Z.; Starodub, N.; Shpyrka, N.; Ramanaviciene, A.; Ramanavicius, A. Analytical, thermodynamical and kinetic characteristics of photoluminescence immunosensor for the determination of Ochratoxin A. Biosens. Bioelectron. 2018, 99, 237–243.
- Viter, R.; Savchuk, M.; Starodub, N.; Balevicius, Z.; Tumenas, S.; Ramanaviciene, A.; Jevdokimovs, D.; Erts, D.; Iatsunskyi, I.; Ramanavicius, A. Photoluminescence immunosensor based on bovine leukemia virus proteins immobilized on the ZnO nanorods. Sens. Actuators B Chem. 2019, 285, 601–606.
- Elledge, S.K.; Zhou, X.X.; Byrnes, J.R.; Martinko, A.J.; Lui, I.; Pance, K.; Lim, S.A.; Glasgow, J.E.; Glasgow, A.A.; Turcios, K.; et al. Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection. medRxiv 2020, preprint.
- Dixon, A.S.; Schwinn, M.K.; Hall, M.P.; Zimmerman, K.; Otto, P.; Lubben, T.H.; Butler, B.L.; Binkowski, B.F.; Machleidt, T.; Kirkland, T.A.; et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 2016, 11, 400–408.
- Kang, S.; Yang, M.; Hong, Z.; Zhang, L.; Huang, Z.; Chen, X.; He, S.; Zhou, Z.; Zhou, Z.; Chen, Q.; et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B 2020, 10, 1228–1238.
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569.
