Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Plant Sciences
The regulation of transcription factors on plants is not single but is regulated by levels of transcription factors at different levels, forming a huge regulatory network and playing a regulatory role. So, what is the regulation between MYB transcription factor and plant secondary wall synthesis? Next, we will try to explain it in detail. In this part, we discuss first the regulation of SCW biosynthesis by MYB46/83 as the second main switch. Next, we consider how other MYB TFs regulate cell-wall biosynthesis in plants.
- secondary cell wall biosynthesis
- MYB transcription factors
- lignification
- classification
- MYB46/83
1. Mechanism by Which MYBs Regulate Lignification
MYB transcription activators/repressors participate in various enzymatic reactions in the phenylpropane metabolic pathway to regulate lignification (Figure 2) [92,93]. Detailed promoter and electrophoretic mobility shift assay of phenylpropane biosynthetic genes, including PAL and 4CL, has shown that the cis-elements corresponding to the MYB TF-binding motif are necessary for coordinated activation of monolignol pathway genes [35,94,95,96,97]. One such element is the AC element (also known as C1-motif, PAL-box, or H-box, divided into I, ACCTACC; II, ACCAACC; and III, ACCTAAC), which is rich in AC sequences. With few exceptions, MYB TFs regulate gene expression by binding to AC elements in the promoter regions of downstream lignin biosynthesis-pathway genes [26,98]. When MYB combines with a specific promoter, the second and third helices form an HTH structure and the third helix functions to directly recognize a particular DNA sequence motif [14]. In SCW biosynthesis, in-depth exploration of the binding mode between MYB transcription factors and AC elements will enable editing of AC elements by genetic engineering to regulate SCW synthesis. However, the mechanisms underlying the selective binding of SCW TFs to the promoters of specific SCW-biosynthesis genes are unclear.
2. MYB46 and MYB83 Are the Second Layer of the Main Switch for Secondary Cell-Wall Biosynthesis
MYB TFs can be activated in multiple ways. Throughout the formation of the secondary wall, the NAC (NAM/ATAF/CUC) TFs acts as the first-level main switch of SCW biosynthesis and activates downstream TFs to regulate the entire SCW biosynthetic network. MYB46/MYB83 act as the second-level main switch of SCW biosynthesis, serving as molecular tools for improving plant biomass (Figure 3) [26,99].
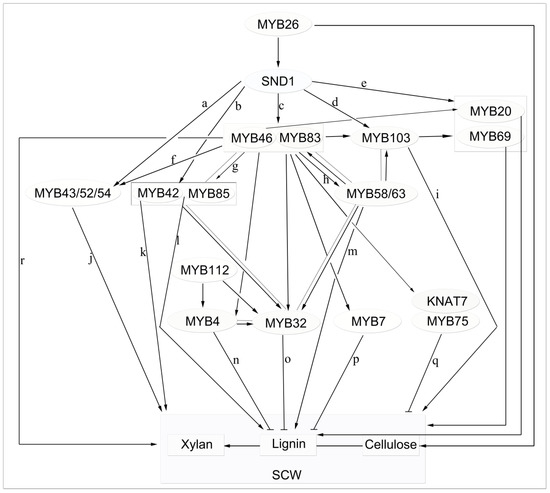
Figure 3. Proposed model of MYB-mediated secondary cell wall (SCW) regulation in Arabidopsis. Solid black and flat-headed arrows represent positive and negative transcriptional regulation, respectively. Dashed lines represent co-expression relationships, MYB26 and SND1, MYB46/83, and other TFs represent the first, second, and third layers of the transcriptional regulatory network, respectively. The expression of 9 SND1-regulated transcription factors, namely, MYB20, MYB42, MYB43, MYB52, MYB54, MYB69, MYB85, MYB103, was developmentally associated with cells undergoing secondary wall thickening (a, b, c, d, e; [72]). MYB46 and MYB83 serve as the second layer of the main switch for secondary cell wall biosynthesis, which activate downstream transcription factors (including MYB20, MYB42, MYB43 and MYB85) by binding to SMRE sequence in an SCW MYB-responsive element (f, g, h, r; [101,103]) and directly or indirectly regulate the biosynthesis of the secondary wall. MYB42, MYB43, MYB85 (j, k, l; [109]), MYB58, MYB63 (m; [110]), MYB103 (i; [37]) are transcriptional regulators that directly activate lignin biosynthesis genes during secondary wall formation in Arabidopsis. MYB4, MYB7, MYB32, MYB75 are inhibitors of lignin biosynthesis (n, o, p, q; [60]). The concerted actions of the MYB TFs in this network leads to a coordinated activation of SCW biosynthetic genes, which results in the synthesis of lignin, cellulose, xylan.
MYB46 and MYB83—two of the earliest discovered lignin-specific TFs—are direct targets of SND1 (secondary wall-associated NAC domain) in Arabidopsis and not only modulate the lignin synthesis pathway but also redundantly activate SCW formation [35,71,72,100]. MYB46 and MYB83 are expressed in vascular tissue and fibers, and their dominant inhibition or RNA interference inhibition markedly suppresses secondary-wall thickening in fibers and vascular tissue leading to collapse of the vascular phenotype. Similar to secondary wall NAC (SWN), overexpression of MYB46 and MYB83 induced ectopic secondary cell wall synthesis [72,100,101]. By analyzing the promoter sequences of downstream genes regulated by MYB46, Zhong et al. found that MYB46 and MYB83 regulate SCW biosynthesis during wood formation by binding to a 7-bp conservative DNA sequence in an SCW MYB-responsive element [SMRE, Secondary wall MYB-responsive element; ACC(A/T)A(A/C)(T/C)] [102,103]. However, the regulation of SCW biosynthesis is more complex than formerly thought. The expression of SCW-biosynthesis genes is regulated by the coordinated actions of multiple MYBs, including activators and repressors [35,104], via binding to not only AC elements (one type of SMRE) but also other SMRE sites. Similar to the promoters of lignin-biosynthesis genes, those of cellulose- and xylan-biosynthesis genes contain multiple SMRE sequences, suggesting that MYBs bind to and activate SMRE sites in the promoters of cellulose- and xylan-biosynthesis genes. Another MYB TF, MYB26, may act as a master switch of SCW biosynthesis in anther endothelial cells—its mutation causes the loss of anther endothelial cell secondary-wall thickening and the anther-dehiscence phenotype. Also, its overexpression leads to ectopic deposition of the secondary wall [105]. MYB26 directly regulates NST1 and NST2, which are critical for inducing secondary thickening biosynthesis genes [106]. The four functional homologous genes MYB TF (PtrMYB2/3/20/21) of MYB46/83 in another model plant poplar, PtrMYB2/3/20/21, are also transcriptional master switches controlling secondary-wall biosynthesis during wood formation that bind to secondary wall NAC-binding element (SNBE) sites in their target gene promoters, thereby activating their expression [107]. Interestingly, the four PtrMYBs exhibit marked differences in how they activate their target genes. One possibility is that they exhibit differential expression patterns in different organs and tissues [108]. Alternatively, they may have different binding affinities for the various SMRE sequences in the promoters of their target genes.
The discovery of the hierarchical transcriptional network that regulates SCW biosynthesis in Arabidopsis was a major breakthrough. However, the regulation of secondary wall formation is more complex than formerly thought, involving positive and negative regulation, dual function regulation, feedback loops, and crosstalk among combinatorial complexes and pathways [103]. Does this affect the transmission of signals related to lignin synthesis by influencing TF-TF, MYB gene-TF, and/or MYB gene-MYB gene interactions? Clarification of the SCW regulatory network warrants further research.
3. Downstream Targets of MYB46/MYB83
3.1. In Arabidopsis
MYB46 and MYB83 activate downstream TF expression [105]. From the metabolic model, MYB46 and MYB83 regulate a series of downstream MYB TFs involved in lignin biosynthesis, including the lignin-activating factors MYB58, MYB63, and MYB85 and the lignin inhibitors MYB4, MYB7, and MYB32 (Figure 3).
Lignin-specific MYBs—MYB58, MYB63, and MYB85—regulate the biosynthesis of lignin rather than cause the deposition of cellulose and hemicellulose (Table 2). Their overexpression leads to activation of lignin-biosynthesis genes and ectopic deposition of lignin in cells that are usually not lignified [35,72,101]. It has long been thought that lignin-specific MYBs bind to AC elements in the promoters of lignin-biosynthesis genes and thereby activate the lignin-biosynthesis pathway [36,43]. MYB58 and MYB63 were first reported as lignin-specific transcriptional activators in Arabidopsis [35]. They have been shown to bind to AC elements and regulate genes involved in lignin biosynthesis (including early genes such as PAL, C4H, and 4CL) but not those involved in cellulose or xylan biosynthesis, which is congruent with the proposed model of regulation of lignin gene expression via AC cis-elements [35]. MYB85 activated the expression of the lignin-biosynthesis gene 4CL1 in a transient assay of Arabidopsis protoplasts (Figure 2) [76].
Table 2. The main MYB transcription factor that regulates secondary wall synthesis in Arabidopsis and poplar.
| Species | MYB TFs | Ortholog in Arabidopsis thaliana | Annotation | References |
|---|---|---|---|---|
| Arabidopsis thaliana | AtMYB3 | inhibit the accumulation of lignin | [111] | |
| AtMYB4 | - | inhibit the accumulation of lignin | [111] | |
| AtMYB7 | - | inhibit the accumulation of lignin | [111] | |
| AtMYB15 | - | promote the synthesis of lignin | [33] | |
| AtMYB20 | - | promotes the accumulation of lignin | [112] | |
| AtMYB32 | - | inhibit the accumulation of lignin | [111] | |
| AtMYB43 | - | promotes the accumulation of lignin | [112] | |
| AtMYB46 | - | promote the synthesis of cellulose, lignin, and hemicellulos | [113] | |
| AtMYBB58 | promotes the accumulation of lignin | [35] | ||
| AtMYB61 | - | promotes the accumulation of lignin | [70] | |
| AtMYB63 | - | promotes the accumulation of lignin | [35] | |
| AtMYB75 | - | inhibit the accumulation of lignin | [114] | |
| AtMYB83 | - | promote the synthesis of cellulose, lignin, and hemicellulos | [113] | |
| AtMYB85 | - | promotes the accumulation of lignin | [72] | |
| AtMYB103 | - | promotes the accumulation of lignin and cellulose | [37] | |
| Poplar | PtrMYB2/3/20/21 | MYB46/83 | promote the synthesis of cellulose, lignin, and hemicellulose | [115] |
| PtrMYB6 | inhibit the accumulation of lignin | [116] | ||
| PtrMYB55 | AtMYB55 | promote the synthesis of lignin and cellulose | [117] | |
| PtrMYB74 | promote the synthesis of cellulose, lignin, and hemicellulose | [118] | ||
| PtoMYB92 | AtMYB85 | promotes the accumulation of lignin, but inhibits the hemicellulose synthesis | [119] | |
| PtrMYB121 | AtMYB55 | promote the synthesis of lignin and cellulose | [117] | |
| PtoMYB125 | AtMYB85 | promotes the accumulation of lignin, but inhibits the hemicellulose synthesis | [119] | |
| PtrMYB152 | AtMYB43 | promotes the accumulation of lignin | [120] | |
| PtoMYB156 | inhibit the accumulation of cellulose, lignin, and hemicellulose | [121] | ||
| PtoMYB170 | AtMYB61 | promotes the accumulation of lignin | [122] | |
| PtrMYB189 | inhibit the accumulation of cellulose, lignin, and hemicellulose | [123] | ||
| PtoMYB216 | AtMYB61 | promotes the accumulation of lignin | [124] | |
| PdMYB221 | inhibit the accumulation of cellulose, lignin, and hemicellulose | [125] |
MYB46, MYB83, and the downstream lignin regulator MYB4 and its homologs MYB7 and MYB32, which belong to subgroup 4 of R2R3-MYB TFs, directly inhibit lignin biosynthesis [62,111,126,127]. The promoter element bound by MYB4 [the 7-bp conserved sequence ACC(A/T)A(A/C)(T/C)] is similar to the SMRE of Arabidopsis. MYB4 regulates the expression of genes related to SCW synthesis by binding to the SMRE sites of downstream target genes or via mitogen-activated protein kinase in A. thaliana and Pinus taeda [26,105]. MYB4, MYB7, and MYB32 have a conserved ethylene-reactive element binding factor-related amphiphilic repression (EAR) motif and GY/FDFLGL motif at the C terminus [62,111]. The GY/FDFLGL motif contributes to the interaction between MYB TFs and SUPER SENSITIVE TO ABA AND DROUGHT 2 (SAD2) [111]. SAD2 is an imported β-like protein that mediates the nuclear translocation of MYB4, MYB7, and MYB32 as well as inhibits the expression of its target genes (e.g., C4H) (Figure 2) [111]. MYB3 is a newly discovered repressor of phenylpropane biosynthesis in A. thaliana and is one of the four members of R2R3-MYB subgroup 4 [62]. The inhibition by MYB3 of C4H expression is directly regulated by the core inhibitors LNK1 and LNK2, which promote the binding of MYB3 to the C4H promoter (Figure 2) [62]. In addition, MYB repressors downregulate AtNST3/SND1 expression in vitro, and AtNST3/SND1 directly regulates AtMYB32 [93]. In view of this, negative feedback of the VNS-MYB network enables fine-tuning of the formation of secondary walls [128]. Except Sg4, members of other subgroups of MYB negatively regulate SCW biosynthesis by interacting with other TFs. For example, the MYB-R3 domain of MYB75 [114] (also known as PAP1) in Arabidopsis and MYB6 [116], MYB26 [106] in transgenic poplar physically interact with the KNOX TF KNAT7, forming a complex that inhibits the development of SCWs in poplar and Arabidopsis. The complex triggers a reduction in deposition and biosynthesis gene expression, which hinders SCW development.
3.2. In Poplar
Most of our understanding of secondary growth comes from the study of Arabidopsis [129]. However, secondary growth in woody perennials is different from that in Arabidopsis roots or hypocotyls [130]. Therefore, identifying the genes that regulate secondary growth in representative woody plant poplar is a top priority [115]. PtrMYB2, PtrMYB3, PtrMYB20, and PtrMYB21 are the functional orthologs of Arabidopsis MYB46 and MYB83, and they regulate poplar secondary-wall biosynthesis by binding to and activating SMRE sequences [105,115]. Like the Arabidopsis SWNs [131,132], PtrWNDs bind to the SNBE sites in the promoters of PtrMYB2/3/20/21 and thereby activate their expression [107]. The findings that these four PtrMYBs all are capable of activating secondary wall biosynthetic genes in poplar trees indicate that these PtrMYBs might function redundantly in regulating secondary wall biosynthesis during wood formation. But why poplar evolved to retain all these four PtrMYBs. One possibility is that although they are all transcriptional activators of secondary wall biosynthesis, they exhibit differential expression patterns in different organs and tissues [108]. Another possibility is that they might differentially activate their target genes as they show differential binding affinity toward different SMRE sequences that are present in promoters of their target genes. Therefore, the expression of these four PtrMYBs might be required for a full strength of transcriptional activation of secondary wall biosynthesis. This is the same as MYB46 and MYB83 in Arabidopsis as the T-DNA knockout mutation of either MYB46 or MYB83 alone does not cause an apparent reduction in secondary wall thickening [71]. Although the functions of some orthologous R2R3-MYB TFs from Arabidopsis and poplar appear to be conserved in regulating SCW biosynthesis, the transcriptional regulation network of SCW biosynthesis may be different in herbaceous and woody plants. Unlike Arabidopsis AtMYB85 which can promote the synthesis of cellulose, lignin, and hemicellulos, its homologues PtoMYB92 and PtoMYB125 can promote the accumulation of lignin but inhibit the synthesis of hemicellulose [119]. Studies have also shown that in the phylogenetic analysis, PtoMYB216 protein groups in the lignification-related R2R3-MYB clade and it is most similar to AtMYB61 from Arabidopsis [124]. AtMYB61 is related to the ectopic lignification of plants [70]. PtoMYB216 is related to the modification of the cell wall of poplar xylem. This may be caused by differences in species [124]. Although the internal MYB transcription factors in plants have different regulation on the secondary wall, they all follow the hierarchical regulation mode of VNSs-MYB-TFs-SCW. Perhaps this can provide a foundation for us to further study the regulation mechanism of the secondary wall.
Similar to Arabidopsis, MYB subgroup 4 members—downstream regulators of PtrMYB2/3/20/21—PtoMYB156 [121], PtrMYB189 [123] and PdMYB221 [125,133,134] are negative regulators of lignin biosynthesis. This is the same as transcription factors such as EgMYB1 [135], BpMYB4 [136], CmMYB8 [137], AmMYB308 [138], ZmMYB42 [139] and ZmMYB31 [104], which are also negative regulators of lignin biosynthesis. Except for PtrMYB189, all of the above-mentioned subgroup 4 members and other MYB repressors have a C-terminally conserved EAR motif, with the expression of these essential genes for repression demonstrated in vitro and in planta [111,112,140]. For PtrMYB189, site-directed deletion and mutagenesis of 13 amino acids (277–289, GDDYGNHGMIKKE) at the C terminus of MYB indicated the importance of this region in target inhibition [123]. Also, numerous MYB TFs enhance cell-wall properties and wood formation. For example, PtrMYB121 directly binds to and activates the promoters of genes related to lignin and cellulose synthesis, thus regulating SCW formation [117]. PtrMYB152, the homolog of the Arabidopsis R2R3-MYB TF AtMYB43, acts as a specific transcriptional activator of lignin biosynthesis during the formation of poplar wood. Overexpression of PtrMYB152 increased the thickness of the secondary wall in plants [120]. PtrMYB92 [119], PtrMYB18, PtrMYB74, PtrMYB75, PtrMYB121, and PtrMYB128 [131] activate the promoters of all three main wood component-biosynthesis genes. In addition, in the third layer, the PtrMYB161 TF binds to multiple sets of target genes, allowing it to act as both an activator and a repressor [141]. It directly regulates the expression of two syringyl-specific monoxylinol genes (PtrCAld5H1 and PtrCAld5H2) [133,142,143] and two key SCW cellulose-synthase genes, PtrCesA4 and PtrCesA18 (PtrCesA8-B) [144,145].
Recent studies have shown that changes in the status of MYB transcription factors can affect the biosynthesis of lignin. For example, phosphorylation of LTF1, an MYB transcription factor in Populus, acts as a sensory switch regulating lignin biosynthesis in wood cells. When LTF1 becomes phosphorylated by PdMPK6 in response to external stimuli such as wounding, it undergoes degradation through a proteasome pathway, resulting in activation of lignification. Expression of a phosphorylation-null mutant version of LTF1 led to stable protein accumulation and persistent attenuation of lignification in wood cells [135]. Moreover, the post-translational regulation of MYB transcription factors, especially their ubiquitination regulation, is closely related to the biosynthesis of lignin. Endoplasmic reticulum-localized E2 ubiquitin-conjugating enzyme 34 (PtoUBC34) interaction with lignin repressors MYB221 and MYB156 regulates the transactivity of the transcription factors in Populus tomentosa. This specific interaction allows for the translocation of TFs PtoMYB221 and PtoMYB156 to the ER and reduces their repression activity in a PtoUBC34 abundance-dependent manner [146]. The above studies show the presence of a complex MYB regulatory network in poplar, similar to that in Arabidopsis, which regulates secondary-wall biosynthesis. Therefore, research on the MYB regulatory networks in Arabidopsis and poplar will enhance the understanding of secondary-wall biosynthesis.
Other aspects of the network require further study, such as the patterns of genetic interaction within the lignin-biosynthesis pathway and how the multigene-coordinated network functions in wood formation. Therefore, plants has a complex transcriptional network that regulates its SCW deposition program, as summarized in Figure 3.
4. Other Elements That Interact with MYB Transcription Factors to Regulate Secondary-Wall Biosynthesis
4.1. Noncoding RNAs
The regulation of secondary walls by noncoding RNAs (ncRNAs), such as microRNAs (miRNAs) and long ncRNAs (lncRNAs), has been a topic of interest. Notably, miRNAs, a class of endogenous ncRNAs consisting of approximately 21–23 nucleotides, play important roles in plant development by cleaving target mRNAs with perfect or near-perfect complementarity [147,148]. The miRNA–MYB network regulates secondary-wall biosynthesis in plants [149] by modulating the activities of enzymes (e.g., CAD and POX) related to phenylpropane metabolic pathways [150]. For example, higher expression of MYBs in MIM858 (an artificial miRNA858 target mimic) lines leads to redirection of the metabolic flux towards the synthesis of flavonoids at the cost of lignin synthesis [149]. Alternatively, miRNAs post-transcriptionally regulate MYB genes related to secondary-wall formation [14,61,151,152]. Lignin biosynthesis is also regulated by coordinated networks involving TFs, miRNAs, and lncRNAs, depending on the genetic effects of the loci [153]. High-throughput RNA sequencing showed that the interaction between lncRNAs, miRNAs, and TFs (including MYBs) contribute to wood formation in Populus. tomentosa [154]. There are few studies on the roles of ncRNAs and MYB TFs in SCW formation. Comparison of differentially expressed miRNA (DEmiRNA) and target gene annotation between poplar and larch suggested the different functions of DEmiRNAs and divergent mechanism in wood formation between two species [155]. To increase our understanding of SCW biosynthesis in plants, these regulatory networks involving TFs, miRNAs, and lncRNAs need to be investigated.
4.2. Plant Hormones
MYB TFs also stimulate plant hormone-mediated plant lignification [58]. For instance, growth hormone, cytokinin, brassinolide and abscisic acid regulate SCW biosynthesis by directly regulating MYB TFs in Arabidopsis, rice, and other plant species [156,157]. ABA has been reported to be involved in the regulation of lignin biosynthetic genes and TF regulators that respond to the lignin accumulation process in plants [158]. For example, ABA induced lignin biosynthesis by promoting the expression of CgMYB58 and its target genes in HR, HB and KP juice sacs [159]. The latest research shows that melatonin can affect the expression of MYB transcription factor, thereby regulating the synthesis of lignin [160]. Also, certain factors combine with hormone-related elements in the MYB promoter region to regulate plant lignification. Auxin response factors (ARFs) are important regulators of lignin biosynthesis in various biological processes in plants. ARF8.4, a flowering-related spliceosome, binds to auxin-related elements in the MYB26 promoter and activates its transcription, thereby controlling interior-wall lignification [161]. Despite these advances, the key plant-hormone-related regulatory nodes in the lignin-biosynthesis pathway have not been elucidated [60]. In-depth exploration of the regulatory network involving MYB TFs and plant hormones will facilitate genetic strategies for increasing plant lignin content.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22073560
This entry is offline, you can click here to edit this entry!
