Normal mammary epithelial cells are heterogeneous and organized in hierarchical fashion, in which the mammary stem cells (MaSC) lie at the apex with regenerative capacity as well as plasticity.
- mammary stem cells
- morphogenesis
- epithelial plasticity
- multipotent
- unipotent
1. Introduction
Normal tissues including the mammary gland consist of a cellular hierarchy; tissue specific (adult) stem cells (SC) at the apex with the ability to self-renew and generate all progeny, committed progenitors with a limited differentiation capacity, and terminally differentiated cells which constitute the functional gland. Adult SCs are distinguished from embryonic stem cells (ESC) in that their ability to generate progeny of distinct cell types is largely restricted to the particular organ from which SC originated. A series of studies, however, have challenged the notion of the traditional lineage restriction of organ specific SCs, demonstrating evidence that trans-differentiation of adult SCs into mature cells of different tissues is possible [1][2]. The discovery of induced pluripotent stem cells (iPSC) from a wide range of differentiated cell types [3][4] and subsequent studies demonstrating direct reprogramming of one adult cell into another functional cell [5][6][7][8][9] provided an indirect support for the possibility of trans-differentiation. Although the mechanism of action of trans-differentiation still remains elusive, it is clear that given the correct series of input signals, a cell can directly be pushed into a different cell type [5][6][7][8][9][10][11]. In normal development and under tissue homeostasis, lineage restriction of any adult stem cells is maintained by a lack of these unusual combinations of signals. However, we are beginning to appreciate that many of the experimental settings (transplant experiments) and pathological conditions (for example, tumor microenvironment) lead to cells experiencing a set of signals that induce a more stem-like state somewhere in between normal adult stem cells and iPSCs. Similarly the phenotypic and functional properties of mammary stem cells (MaSC) in mammary gland development are under intense investigation. Extensive studies thus far have proposed two opposing models of MaSCs. Majority of these studies have concluded that bipotent stem cells are able to give rise to both luminal and basal populations [12][13][14][15][16][17][18][19][20][21]. However, other studies proposed that the adult mammary gland only contains unipotent stem cells, each of which generate only luminal or basal lineages [20][21][22][23][24]. What these stem cells have in common though is that they have been shown to cross–communicate with their microenvironment to maintain homeostasis, which ensures the generation of mature functional cells throughout the life of organism without depletion of stem cell pools [12][25][26][27][28].
2. Long-Lived Quiescent MaSCs Which Are Activated in Response to Stimuli
Although both lineage tracing and transplantation assays supported the existence of bipotent embryonic MaSCs, whether these bipotent embryonic stem cells are maintained in adult mammary gland remains controversial [19][20][29]. Recent studies have explored the idea of quiescent stem cells as discovered in other systems such as hematopoietic system [30], and reported the existence of long-lived, quiescent MaSCs which re-enter the cell cycle and give rise to both lineages in response to stimuli in adult mammary gland [12][13][29][31]. A fluorescent dye, PKH-26 which binds to cell membranes and segregates in daughter cells in each cell division, was used to determine the rate of cell division in mammosphere forming basal cells and repopulating efficiencies in transplantation assays [31]. Interestingly, only slow-cycling PKHhi cells (0.5–1%), sorted from secondary mammospheres, were able to reconstitute mammary gland, while no mammary reconstitution was observed in mice transplanted with PKH+ or PKHlo cells isolated similarly from secondary mammospheres [31]. Two subsequent studies corroborated these findings, supporting the evidence of quiescent MaSCs becoming activated in adult mammary glands upon stimuli. In an effort to characterize basal compartment, Fu et al. performed gene expression profiling of Lgr5+ cells and identified Tetraspanin8 (Tspan8) as a surface marker [12]. Fractionation based on the Lgr5 and Tspan8 expressions (Lgr5+Tspan8hi, Lgr5−Tspan8hi, Lgr5+Tspan8− and Lgr5−Tspan8−) revealed that although all subsets displayed varying degrees of clonogenic potential in vitro and repopulating capacity in transplantation assays, ductal outgrowths were not identical. Lgr5+Tspan8hi cell population not only exhibited a superior repopulating capacity, but also gave rise to all lineages. Interestingly, Lgr5+Tspan8hi subset consisted of slow cycling cells with a distinct epigenetic profile [12]. Furthermore, these otherwise quiescent MaSCs were shown to be activated by steroid hormones as evidenced by reduced proportion of quiescent (Lgr5+Tspan8hi) subset in mammary glands of pregnant mice compared to those of virgin mice [12]. Together the data reveal the existence of quiescent MaSC subsets which may be activated upon stimuli in adult mammary glands (Figure 1).
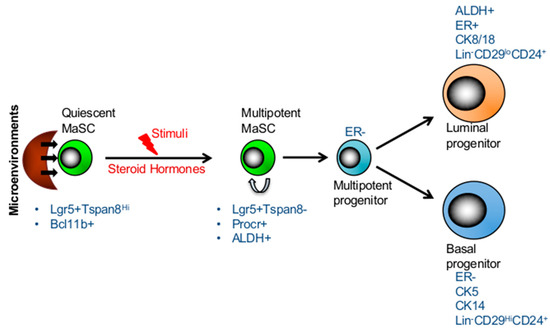
Figure 1. Hierarchical structure and plasticity of mammary stem cells. The transition between long-lived, quiescent, and proliferating MaSCs is regulated by its microenvironment. Quiescent MaSCs can re-ender the cell cycle and give rise to both lineages in response to stimuli, such as steroid hormones in the adult mammary gland. The markers of two subsets of stem cells and progenitors are presented in the figure.
Employing single cell gene expression analyses, Cai et al. identified a quiescent cell population which expressed high levels of Bcl11b and was restricted to the basal compartment [13]. The clonogenicity and transplantation assays of Bcl11bhi and Bcl11blow subsets within the basal Lin-CD49fhiCD24med compartment, demonstrated that Bcl11bhi subset was substantially more efficient in generating colonies and engraftment capacity. Furthermore, conditional deletion of Bcl11b in mammary gland resulted in significant reductions of postnatal mammary gland development due to exhaustion of ductal mammary epithelium [13]. When compared with Procrhi cell subset [14], Bcl11b marked a distinct cell population within the basal compartment with CD49fhiCD24med phenotype [13]. Whereas Procr+ cells were actively cycling, Bcl11b+ cells were mainly quiescent and that this gene functionally regulated the homeostasis of quiescent basal cell population [13]. Although quiescent and cycling MaSCs in human mammary glands has not been well defined, so far the studies have established the EpCAM−CD49fhi phenotype being the marker of human MaSCs [32][33][34]. In addition human multipotent and basal progenitors expressed surface markers such as CD10, CD90 and TP63 [32][34]. Besides these markers, the expression of enzymatic activity of ALDH1 has also been shown to label both human and mouse multipotent MaSC and luminal progentors [35][36][37][38]. Furthermore, studies established that both mouse and human MaSCs and basal progenitors lack ER expression (ER-) while luminal progenitors show low levels of ER (ER+/−) expression (Figure 1) [33][34][39]. However, in mouse mammary gland CD49fhiSca-1+ population was primarily ER-positive (ER+) as previously shown [40]. Together these studies provide a strong evidence for hierarchical organization of the mammary gland and the quiescent MaSCs at the apex which are important for repopulating and long-term maintenance of the mammary gland.
3. Mammary Stem Cell Plasticity Regulated by the Microenvironment during Mammary Development
Epithelia in general show enormous heterogeneity and functional plasticity throughout the body [25]. Although mammary epithelium is the functional unit of the mammary gland, its development and maintenance also requires a complex cross-talk with the surrounding stroma [26]. Understanding this cross-talk has been proven to be a challenging task due to complexities of the microenvironment in which mammary epithelia go through morphogenesis, lactation and involution in each cycle of pregnancy. One such example of stroma contributing the SC plasticity is demonstrated in hair regeneration. In resting hair follicles, the bulge is comprised of heterogeneous stem cell population, while CD34 expression marks quiescent SCs in both upper and lower bulge, Lgr5 expressing and actively cycling SCs are restricted to the lower bulge [41]. A recent study reported that the elimination of the cycling Lgr5+ cells by using diphtheria-toxin-mediated cell ablation abrogated the hair regeneration but it was reversed during the recovery phase by activation of quiescent CD34+ stem cells via the inflammatory responses [42]. The fact that the activation of otherwise quiescent CD34+ stem cells by inflammatory responses provides a compelling evidence for the role of the microenvironment in regulating stem cell plasticity and tissue homeostasis. In line with these studies, it has been suggested that inflammatory responses and microenvironmental cues may lead to stem cell plasticity leading to heterogeneous MaSC phenotypes [43]. Interestingly, when FACS-sorted YFP+ basal and luminal cells were co-transplanted, only basal cells gave rise to both basal and luminal lineages. In order to explain the discrepancy of the data, Ven Keymeulen and colleagues argued that the experimental setting of co-transplantation forces this multi-lineage differentiation of basal cells [22]. In line with the notion, although it is yet to be experimentally determined, it may entirely be possible that transplantation assay may potentially induce inflammatory responses which in turn activate otherwise quiescent MaSCs [12][13] thereby regenerating the mammary gland. In addition, emerging studies implicate a secondary advantage of the MaSC plasticity or remaining quiescent that provides protection from pathological and immunological insults [44][45]. Type I interferons (IFN-1) induces a transient HSC proliferation in acute inflammation, however, in response to chronic IFN-1 exposure, HSCs rapidly return to quiescence [45]. This reestablished quiescence protects HSCs from IFN-1-induced exhaustion unless forced back into the cell cycle due to in vitro culture or transplantation conditions [45]. A similar mechanism was reported to protect normal and malignant mammary stem cells from the cytotoxic effect of IFN via miR-199a-mediated repression of nuclear receptor corepressor LCOR [44]. It was demonstrated that elevated miR-199 expression and subsequent LCOR repression protects stem cells from differentiation and senescence induced by IFNs that are produced by epithelial and immune cells. Consistent with the notion, quiescent MaSCs located at the proximal region of ductal tree were protected in Lgr5-GFP-IREScreERT2/R26R-tdTomato mice during involution [12].
Although there is yet to be a consensus on the location of quiescent MaSCs in the mammary ductal tree [12][13][46], a distinct location and surrounding microenvironment may also play a role in maintaining them in quiescence [12] (Figure 2).
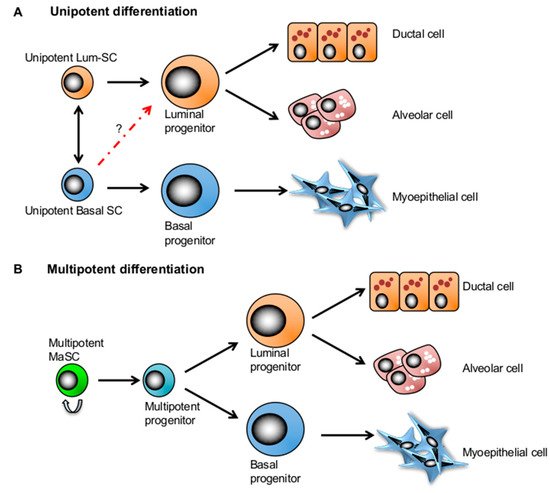
Figure 2. Two competing mammary stem cell (MaSC) models in mammary gland development. (A) The lineage-restricted unipotent MaSC model postulates that each compartment is maintained by its own unipotent stem cells in the adult mammary gland. (B) The bipotent MaSC model proposes that there exist a multipotent stem cell population that can give rise to all lineages of the mammary tissue.
This entry is adapted from the peer-reviewed paper 10.3390/ijms20092357
References
- Merrell, A.J.; Stanger, B.Z. Adult cell plasticity in vivo: De-differentiation and transdifferentiation are back in style. Nat. Rev. Mol. Cell Biol. 2016, 17, 413–425.
- Morrison, S.J. Stem cell potential: Can anything make anything? Curr. Biol. 2001, 11, R7–R9.
- Okita, K.; Nakagawa, M.; Hyenjong, H.; Ichisaka, T.; Yamanaka, S. Generation of mouse induced pluripotent stem cells without viral vectors. Science 2008, 322, 949–953.
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676.
- Chen, Y.; Yang, Z.; Zhao, Z.A.; Shen, Z. Direct reprogramming of fibroblasts into cardiomyocytes. Stem Cell Res. Ther. 2017, 8, 118.
- Hou, S.; Lu, P. Direct reprogramming of somatic cells into neural stem cells or neurons for neurological disorders. Neural. Regen. Res. 2016, 11, 28–31.
- Kelaini, S.; Cochrane, A.; Margariti, A. Direct reprogramming of adult cells: Avoiding the pluripotent state. Stem Cells Cloning 2014, 7, 19–29.
- Ambasudhan, R.; Talantova, M.; Coleman, R.; Yuan, X.; Zhu, S.; Lipton, S.A.; Ding, S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell 2011, 9, 113–118.
- Zhou, Q.; Brown, J.; Kanarek, A.; Rajagopal, J.; Melton, D.A. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 2008, 455, 627–632.
- Rackham, O.J.; Firas, J.; Fang, H.; Oates, M.E.; Holmes, M.L.; Knaupp, A.S.; Consortium, F.; Suzuki, H.; Nefzger, C.M.; Daub, C.O.; et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016, 48, 331–335.
- Rajapakse, I.; Groudine, M.; Mesbahi, M. Dynamics and control of state-dependent networks for probing genomic organization. Proc. Natl. Acad. Sci. USA 2011, 108, 17257–17262.
- Fu, N.Y.; Rios, A.C.; Pal, B.; Law, C.W.; Jamieson, P.; Liu, R.; Vaillant, F.; Jackling, F.; Liu, K.H.; Smyth, G.K.; et al. Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nat. Cell Biol. 2017, 19, 164–176.
- Cai, S.; Kalisky, T.; Sahoo, D.; Dalerba, P.; Feng, W.; Lin, Y.; Qian, D.; Kong, A.; Yu, J.; Wang, F.; et al. A quiescent bcl11b high stem cell population is required for maintenance of the mammary gland. Cell Stem Cell 2017, 20, 247–260.
- Wang, D.; Cai, C.; Dong, X.; Yu, Q.C.; Zhang, X.O.; Yang, L.; Zeng, Y.A. Identification of multipotent mammary stem cells by protein c receptor expression. Nature 2015, 517, 81–84.
- Rios, A.C.; Fu, N.Y.; Lindeman, G.J.; Visvader, J.E. In situ identification of bipotent stem cells in the mammary gland. Nature 2014, 506, 322–327.
- Shackleton, M.; Vaillant, F.; Simpson, K.J.; Stingl, J.; Smyth, G.K.; Asselin-Labat, M.L.; Wu, L.; Lindeman, G.J.; Visvader, J.E. Generation of a functional mammary gland from a single stem cell. Nature 2006, 439, 84–88.
- Kordon, E.C.; Smith, G.H. An entire functional mammary gland may comprise the progeny from a single cell. Development 1998, 125, 1921–1930.
- Stingl, J.; Eirew, P.; Ricketson, I.; Shackleton, M.; Vaillant, F.; Choi, D.; Li, H.I.; Eaves, C.J. Purification and unique properties of mammary epithelial stem cells. Nature 2006, 439, 993–997.
- Makarem, M.; Kannan, N.; Nguyen, L.V.; Knapp, D.J.; Balani, S.; Prater, M.D.; Stingl, J.; Raouf, A.; Nemirovsky, O.; Eirew, P.; et al. Developmental changes in the in vitro activated regenerative activity of primitive mammary epithelial cells. PLoS Biol. 2013, 11, e1001630.
- Spike, B.T.; Kelber, J.A.; Booker, E.; Kalathur, M.; Rodewald, R.; Lipianskaya, J.; La, J.; He, M.; Wright, T.; Klemke, R.; et al. Cripto/grp78 signaling maintains fetal and adult mammary stem cells ex vivo. Stem Cell Rep. 2014, 2, 427–439.
- Nguyen, L.V.; Makarem, M.; Carles, A.; Moksa, M.; Kannan, N.; Pandoh, P.; Eirew, P.; Osako, T.; Kardel, M.; Cheung, A.M.; et al. Clonal analysis via barcoding reveals diverse growth and differentiation of transplanted mouse and human mammary stem cells. Cell Stem Cell 2014, 14, 253–263.
- Van Keymeulen, A.; Rocha, A.S.; Ousset, M.; Beck, B.; Bouvencourt, G.; Rock, J.; Sharma, N.; Dekoninck, S.; Blanpain, C. Distinct stem cells contribute to mammary gland development and maintenance. Nature 2011, 479, 189–193.
- Wuidart, A.; Sifrim, A.; Fioramonti, M.; Matsumura, S.; Brisebarre, A.; Brown, D.; Centonze, A.; Dannau, A.; Dubois, C.; Van Keymeulen, A.; et al. Early lineage segregation of multipotent embryonic mammary gland progenitors. Nat. Cell Biol. 2018, 20, 666–676.
- Lilja, A.M.; Rodilla, V.; Huyghe, M.; Hannezo, E.; Landragin, C.; Renaud, O.; Leroy, O.; Rulands, S.; Simons, B.D.; Fre, S. Clonal analysis of notch1-expressing cells reveals the existence of unipotent stem cells that retain long-term plasticity in the embryonic mammary gland. Nat. Cell Biol. 2018, 20, 677–687.
- Donati, G.; Watt, F.M. Stem cell heterogeneity and plasticity in epithelia. Cell Stem Cell 2015, 16, 465–476.
- Parmar, H.; Cunha, G.R. Epithelial-stromal interactions in the mouse and human mammary gland in vivo. Endocr. Relat. Cancer 2004, 11, 437–458.
- Ingthorsson, S.; Briem, E.; Bergthorsson, J.T.; Gudjonsson, T. Epithelial plasticity during human breast morphogenesis and cancer progression. J. Mammary Gland. Biol. Neoplasia 2016, 21, 139–148.
- Landskroner-Eiger, S.; Park, J.; Israel, D.; Pollard, J.W.; Scherer, P.E. Morphogenesis of the developing mammary gland: Stage-dependent impact of adipocytes. Dev. Biol. 2010, 344, 968–978.
- Boras-Granic, K.; Dann, P.; Wysolmerski, J.J. Embryonic cells contribute directly to the quiescent stem cell population in the adult mouse mammary gland. Breast Cancer Res. 2014, 16, 487.
- Baldridge, M.T.; King, K.Y.; Boles, N.C.; Weksberg, D.C.; Goodell, M.A. Quiescent haematopoietic stem cells are activated by ifn-gamma in response to chronic infection. Nature 2010, 465, 793–797.
- Cicalese, A.; Bonizzi, G.; Pasi, C.E.; Faretta, M.; Ronzoni, S.; Giulini, B.; Brisken, C.; Minucci, S.; Di Fiore, P.P.; Pelicci, P.G. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 2009, 138, 1083–1095.
- Pellacani, D.; Tan, S.; Lefort, S.; Eaves, C.J. Transcriptional regulation of normal human mammary cell heterogeneity and its perturbation in breast cancer. EMBO J. 2019.
- Visvader, J.E.; Stingl, J. Mammary stem cells and the differentiation hierarchy: Current status and perspectives. Genes Dev. 2014, 28, 1143–1158.
- Oakes, S.R.; Gallego-Ortega, D.; Ormandy, C.J. The mammary cellular hierarchy and breast cancer. Cell Mol. Life Sci. 2014, 71, 4301–4324.
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. Aldh1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567.
- Domenici, G.; Aurrekoetxea-Rodriguez, I.; Simoes, B.M.; Rabano, M.; Lee, S.Y.; Millan, J.S.; Comaills, V.; Oliemuller, E.; Lopez-Ruiz, J.A.; Zabalza, I.; et al. A sox2-sox9 signalling axis maintains human breast luminal progenitor and breast cancer stem cells. Oncogene 2019, 38, 3151–3169.
- Shehata, M.; Teschendorff, A.; Sharp, G.; Novcic, N.; Russell, I.A.; Avril, S.; Prater, M.; Eirew, P.; Caldas, C.; Watson, C.J.; et al. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012, 14, R134.
- Eirew, P.; Kannan, N.; Knapp, D.J.; Vaillant, F.; Emerman, J.T.; Lindeman, G.J.; Visvader, J.E.; Eaves, C.J. Aldehyde dehydrogenase activity is a biomarker of primitive normal human mammary luminal cells. Stem Cells 2012, 30, 344–348.
- Lim, E.; Vaillant, F.; Wu, D.; Forrest, N.C.; Pal, B.; Hart, A.H.; Asselin-Labat, M.L.; Gyorki, D.E.; Ward, T.; Partanen, A.; et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in brca1 mutation carriers. Nat. Med. 2009, 15, 907–913.
- Rodilla, V.; Dasti, A.; Huyghe, M.; Lafkas, D.; Laurent, C.; Reyal, F.; Fre, S. Luminal progenitors restrict their lineage potential during mammary gland development. PLoS Biol. 2015, 13, e1002069.
- Jaks, V.; Barker, N.; Kasper, M.; van Es, J.H.; Snippert, H.J.; Clevers, H.; Toftgard, R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 2008, 40, 1291–1299.
- Hoeck, J.D.; Biehs, B.; Kurtova, A.V.; Kljavin, N.M.; de Sousa, E.M.F.; Alicke, B.; Koeppen, H.; Modrusan, Z.; Piskol, R.; de Sauvage, F.J. Stem cell plasticity enables hair regeneration following lgr5+ cell loss. Nat. Cell Biol. 2017, 19, 666–676.
- Wahl, G.M.; Spike, B.T. Cell state plasticity, stem cells, emt, and the generation of intra-tumoral heterogeneity. NPJ Breast Cancer 2017, 3, 14.
- Celia-Terrassa, T.; Liu, D.D.; Choudhury, A.; Hang, X.; Wei, Y.; Zamalloa, J.; Alfaro-Aco, R.; Chakrabarti, R.; Jiang, Y.Z.; Koh, B.I.; et al. Normal and cancerous mammary stem cells evade interferon-induced constraint through the mir-199a-lcor axis. Nat. Cell Biol. 2017, 19, 711–723.
- Pietras, E.M.; Lakshminarasimhan, R.; Techner, J.M.; Fong, S.; Flach, J.; Binnewies, M.; Passegue, E. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type i interferons. J. Exp. Med. 2014, 211, 245–262.
- Scheele, C.L.; Hannezo, E.; Muraro, M.J.; Zomer, A.; Langedijk, N.S.; van Oudenaarden, A.; Simons, B.D.; van Rheenen, J. Identity and dynamics of mammary stem cells during branching morphogenesis. Nature 2017, 542, 313–317.
