Viruses have evolved different strategies to hijack subcellular organelles during their life cycle to produce robust infectious progeny. Successful viral reproduction requires the precise assembly of progeny virions from viral genomes, structural proteins, and membrane components. Such spatial and temporal separation of assembly reactions depends on accurate coordination among intracellular compartmentalization in multiple organelles. Virus trigger the rearrangement and morphology remodeling of intracellular organelles, including the quality control of intracellular organelles, the hijacking of the modified organelle membranes, morphology remodeling for viral replication, and degradation of intracellular organelles by virus-triggered selective autophagy.
- virus
- intracellular organelles
- rearrangement
- remodeling
Rearrangement of intracellular organelles during viral infections
To maximize their viral replication and evade host antiviral responses, viruses have evolved a plethora of strategies to hijack cellular organelles [1-3]. Each step of viral replication is closely accompanied by the rearrangement of intracellular organelles.
Remodeling of the mitochondria for viral replication
The mitochondria are highly dynamic organelles and form interconnected tubular networks, undergoing a balance between fusion and fission in response to intracellular and/or extracellular stresses [4] (Figures 1A and 1B). Mitochondrial fusion involves two sets of key GTPase proteins in mammals: the mitofusin GTPases (Mfns) (Mfn1 and Mfn2) of the outer mitochondrial membrane (OMM) and optic atrophy 1 (OPA1) of the inner mitochondrial membrane (IMM) [5-9]. The Mfns mediate OMM fusion and cristae integrity [10]. However, the OPA1 mediates IMM fusion and cristae integrity by regulating of the mRNA splicing forms, membrane potential, and the adenosine triphosphate (ATP)-dependent diverse cellular proteases [6-8]. Subsequently, OMM fusions are followed by IMM fusion processes, resulting in the concomitant mixing of the mitochondrial contents and merging of two individual mitochondria. In a previous study, Cipolat, S et al. identified that OPA1 specific functional cross-talk with Mfn1 rather than Mfn2 is involved in the mitochondrial fusion of OMM [11]. Mitochondrial fission is a complex process which includes two distinct steps: an initial constriction of mitochondrial membranes and membrane scission. The initial constriction step narrows the mitochondrial tube diameter at the ER-mitochondria intersection zones where ER tubules wrap around the OMM. Manor, U. et al. suggested that actin-nucleating protein spire 1C localizes to the mitochondria, and directly links the mitochondria to the actin cytoskeleton and the ER, and finally promotes actin polymerization at the ER-mitochondria intersections [12]. The membrane scission of the mitochondria is primarily regulated by dynamic relative GTPase protein (DRP-1) [13]. The mitochondrial localization of DRP-1 is a cytosolic factor promoting mitochondrial fission, which powers the constriction and division of the mitochondria primarily through post-translational modification (e.g., phosphorylation) (reviewed by Lee. H et al. [14]). Recent studies have reported that the recruitment of DRP-1 in mammalian cells requires several accessory proteins, such as the mitochondrial fission protein 1 (Fis-1) and mitochondrial fission factor (Mff) [15]. Although such proteins are proposed to constitute the fission complex of the mitochondria, mediating mitochondrial fission using this complex has remained unclear.
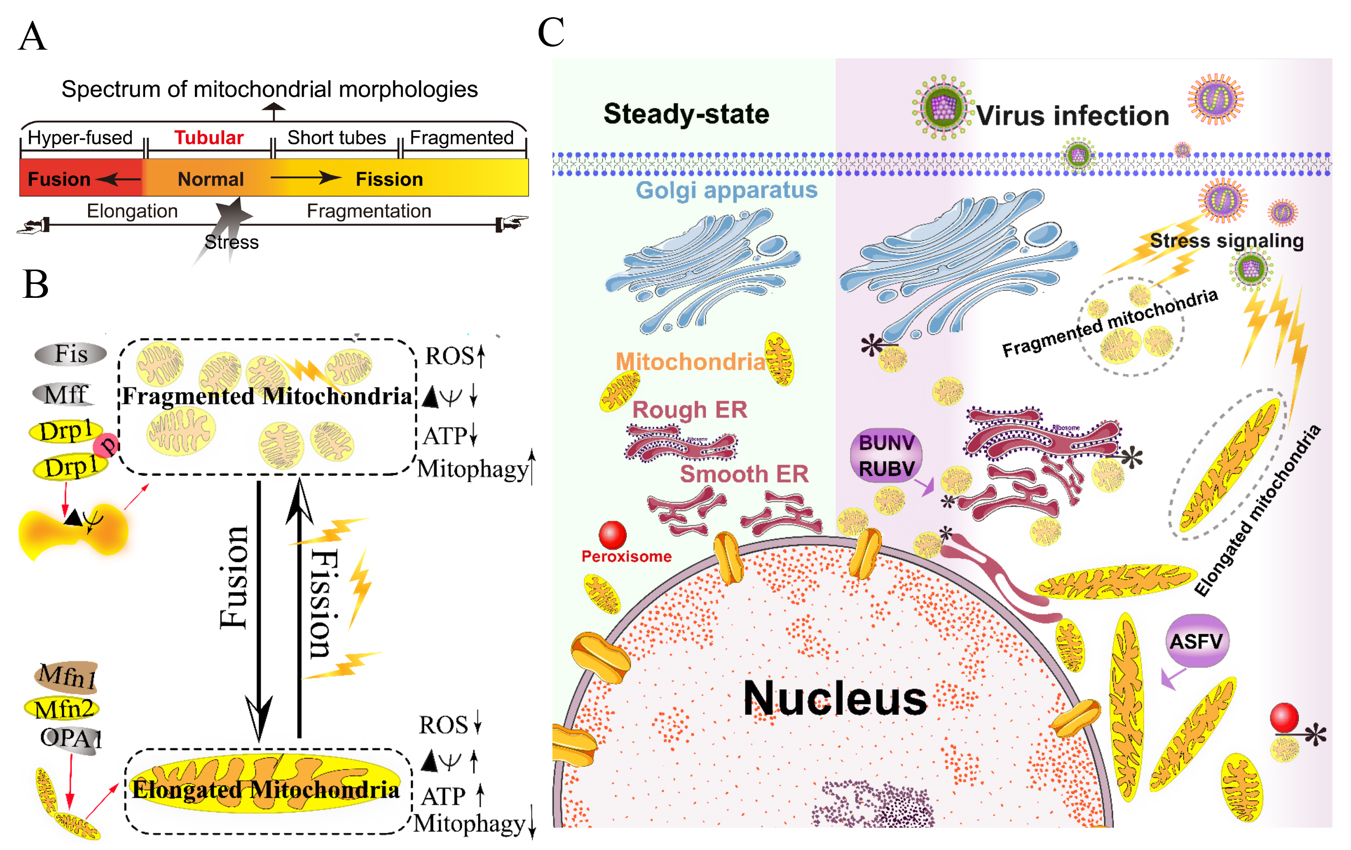
Viruses have evolved several strategies to remodel the mitochondria for viral replication and assembly, including spatial distribution, morphology remodeling, and metabolism reprogramming. To maximize the effectiveness of DNA replication, African swine fever virus (ASFV) infection recruits the mitochondria around the viral factories, associated with the morphology change and accumulation of the mitochondria. It was speculated that the translation and ATP synthesis are coupled and compartmentalized around viral factories to promote virus replication [16] (Figure 1C). Normal mitochondria are dynamic organelles, and form interconnected tubular networks [4, 17] (Figure 1A). The cristae remodeling of the IMM determines the assembly and stability of respiratory chain supercomplexes and respiratory efficiency [18]. In general, the mitochondrial elongation process is associated with the dimerization and activation of the ATPase function to produce additional energy [17, 19]. NDV induces the hyper-fusion of the mitochondria in infected A549 cells (unpublished data), which is similar to the characteristic of Dengue virus (DENV) [20] and severe acute respiratory syndrome-coronavirus (SARS-CoVs) [21]. Notably, except for vaccinia virus (VV) [22], most viruses exploit aerobic glycolysis of the mitochondria for the production of viral progeny [2].
Moreover, viral infections may increase the inter-organellar interactions of the mitochondria with other organelles for replication. Rubella virus (RUBV) [23] and Bunyamwera virus (BUNV) [24] infections increase the membrane interactions among mitochondria, ER, and Golgi (Figure 1C), which is consistent with that of the ER-mitochondria contract that serves as a platform for inter-organellar communication [25].
Figure 1. Morphology remodeling and spatial redistribution of mitochondria triggered by virus infections. (A) The morphological diagram of the mitochondria. Mitochondria form a dynamic network pool, which constantly undergoes rearrangement and turnover. The equilibrium regulation of mitochondrial fusion–fission is essential to maintain the integrity of mitochondria [26]. The morphology of mitochondria was divided into hyper-fused (elongated), tubular (normal), short tubes, and fragmented [17]. (B) Regulation of mitochondrial fusion and fission. Mitochondrial fusion is mediated by mitofusin GTPases MFN1 and MFN2 at the outer mitochondrial membrane (OMM), and OPA1 at the inner mitochondrial membrane (IMM). Mitochondrial fission is driven by the fission machinery complex, which consists of DRP-1, Fis1, and MFF. Mitochondrial hyper-fusion is a pro-survival type, which can increase the ATP production and membrane potential (Δψm), and decrease reactive oxygen species (ROS) and mitophagy [17, 26]. (C) Proposed model for the nuclear aggregation of mitochondria and the possible interplay among intracellular organelles in response to virus infections. * symbol indicates the possible interaction site. Representative virus that increases the interactions among intracellular organelles is shown with purple rectangle. African swine fever virus, ASFV; Rubella virus, RUBV; Bunyamwera virus, BUNV.
To date, several reports have argued the role of Mfns in innate immunity [27-29]. The interaction of Mfns with the adaptor mitochondrial antiviral signaling protein (MAVS) (also called IPS-1, Cardif, or VISA) at the mitochondrial associated membrane (MAM) leads the initiation of the IFN signaling pathway [30, 31]. Meanwhile, MAVS was also reported to interact with MFN2, which leads to the inhibition of inflammatory cytokine production, suggesting the MAM plays a complex role in the regulation of innate immunity [28] (detailed in review [31]). Castanier, C et al. also identified the cross-modulation relationship between mitochondrial dynamic and retinoic acid-inducible gene I protein (RIG-I) like receptor (RLR) signaling activation [27]. Certain viruses, such as influenza A virus (IAV) [32], measles virus (MV) [33], hepatitis B virus (HBV) [34-37], and hepatitis C virus (HCV) [34], induce selective autophagy to degrade fragmented mitochondria and evade innate immunity. Meanwhile, the non-structural (NS) protein 4B of DENV induces mitochondrial elongation via inactivation of DRP-1 and dampens the activation of RLR signal pathway to promote replication [20]. Similarly, the open reading frame-9b (ORF-9b) encoded by SARS-CoVs causes mitochondrial elongation via triggering DRP-1 degradation, and inhibits RLR signaling [21].
Collectively, viruses have evolved several strategies to hijack the mitochondria for viral genome replication and assembly, including the remodeling of mitochondrial morphology and distribution, the regulation of the fusion–fission machinery complex, and the synthesis of ATP production.
Rearrangement of ER and unfolded protein response (UPR) during viral infection
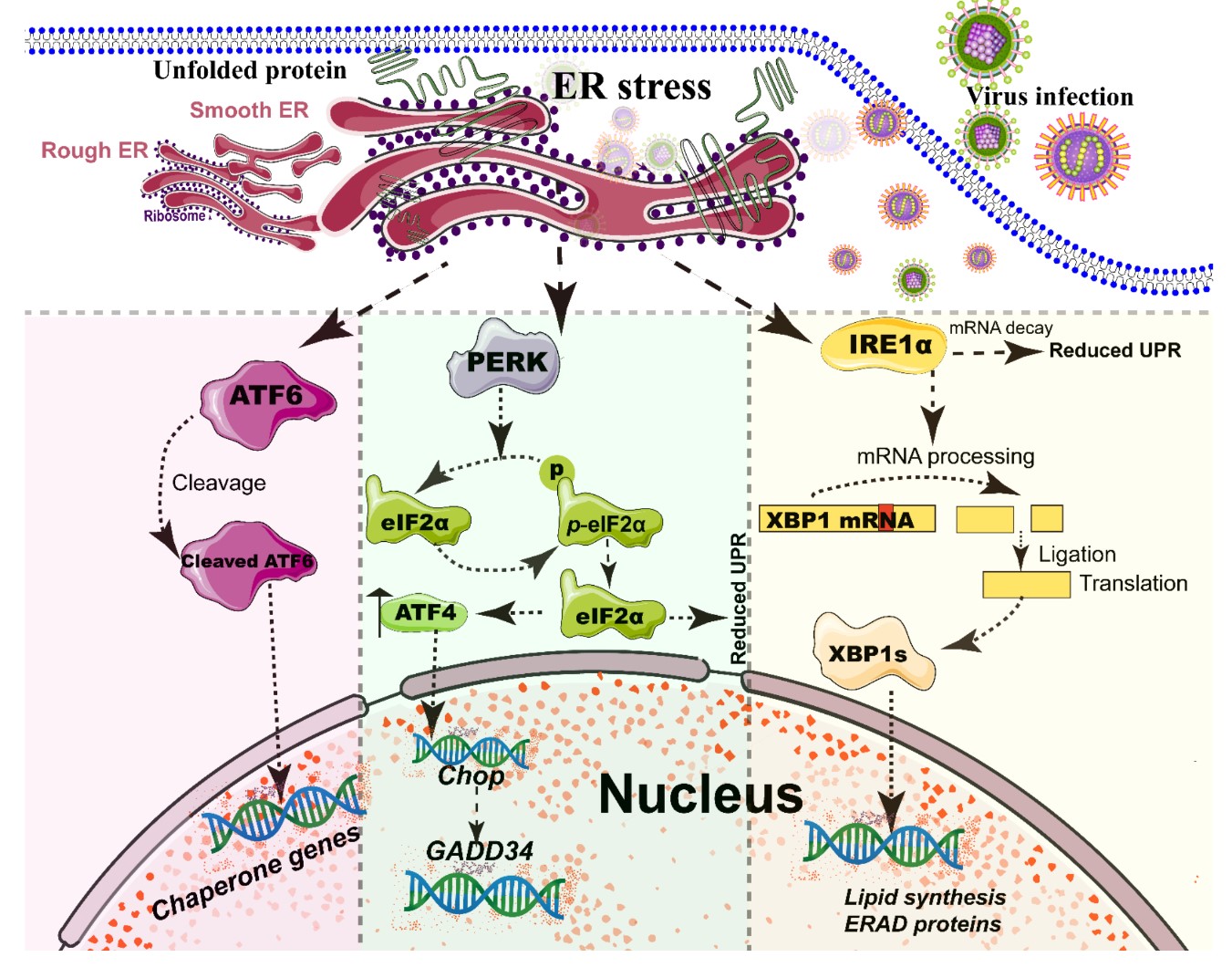
The ER, a single continuous membrane, consists of two primary structural subdomains: the nuclear envelope and the peripheral ER (a polygonal network) [38]. The nuclear envelope of ER consists of two flat membrane bilayers; the peripheral ER is composed of membrane cisternae and dynamic interconnected tubules [38, 39]. The ER is the largest intracellular endomembrane system and has multiple complex functions, including Ca2+ storage, fatty acid synthesis, ion homeostasis, and, in particular, the quality control of newly synthesized proteins [40]. The accumulation of misfolded or unfolded proteins in the ER lumen is known as ER stress [41]. UPR and ER-associated degradation (ERAD) signaling are central to maintain the quality control of the ER [41, 42]. The UPR is a signaling cascade aimed at eliminating misfolding proteins and increasing folding capacity in lumen [41]. The protein-folding conditions in the ER lumen is primarily sensed by three integrated signaling transducers: activating transcription factor 6 (ATF6) [43], double-stranded RNA-activated protein kinase-like kinase (PERK), and inositol requiring enzyme 1α (IRE1α) [25, 44] (Figure 2). Each branch uses a distinct mechanism to drive the transcription of UPR signal transduction, such as ATF6 by regulated proteolysis, PERK by translational control, and IRE1 by non-conventional mRNA splicing [44]. By contrast, ERAD recognizes misfolded proteins and retro-translocates such proteins into the cytoplasm for degradation by the ubiquitin-proteasome-dependent ERAD and the autophagy-lysosome dependent ERAD [42, 45].
A series of studies has reported that viral infections reshape the morphology and membrane remodeling of ER [3, 38], and exploit various strategies to hijack the three branches of UPR for viral replication (Figure 2 and Table 1). The possible explanations were summarized as follows: First, the large malleable surface area of ER is used as a physical scaffold to protect viral RNA from degradation by cellular mRNA decay machinery [40, 46]. RNA viruses have evolved several strategies to avoid the cellular mRNA decay machinery [46]. Second, viruses, particularly most RNA viruses, remodel the ER membrane to form a variety of structures for infectious progeny [47], including single-membrane spherule vesicles, double-membrane vesicles, convoluted membranes, and single-membrane sheets in the ER lumen [38]. Tenorio, R et al. identified that δNS and μNS of reovirus caused tubulation and fragmentation of the ER, respectively, to re-build replication sites [48], indicating that viral proteins play different roles in the rearrangement of ER membranes. Similarly, the NS4A of DENV induces the membrane arrangement of ER lumen in a 2K-regulated manner [49]. Third, viruses recruit the ER membranes into the replication and assembly compartments. The viral cytoplasmic replication site of VV [50, 51], equine arteritis virus (EAV) [52], and polivirus (PV) [53] is derived directly from the ER membrane. Meanwhile, ASFV structural protein p54 plays an important role in the recruitment and transformation of the ER membranes into the envelope precursors [54]. Fourth, viruses increase the capacity and spatial rearrangement to increase ER biogenesis, including membrane protein synthesis, fatty acid change, and Ca2+ storage [40]. For enveloped viruses, the key molecular chaperone of ER, including Bip/GRP78 and calnexin/calreticulin, assists the folding of the extracellular domains of viral membrane glycoproteins, such as GP2a of PRRSV [55], and hemagglutinin-neuraminidase (HN) and fusion (F) proteins of NDV [56], when they translocate into the lumen of the ER. Meanwhile, the reprograming of ER biogenesis, such as Ca2+ storage, is required for viral replication, including HCV [57] and ASFV [58]. Fifth, viruses co-opt or subvert the ERAD processes to re-establish ER homeostasis, which actively exports the malformed proteins from the ER for degradation. Human cytomegalovirus (HCMV) [59] and IAV [60] exploit the ERAD pathway to benefit viral replication. Finally, the membrane remodeling of ER may suppress the activation of host immunity. Upon viral infections, particularly DNA viruses, stimulator of interferon genes (STING), an activated ER adaptor of the cyclic GMP-AMP synthase (cGAS)-STING signaling pathway, translocates from the ER to the ER-Golgi-intermediate compartment (ERGIC) and the Golgi apparatus, and then activates downstream molecules [61-63]. Therefore, we speculate that the morphology remodeling and membrane modification of ER induced by viruses may be involved in the regulation of STING trafficking, EARD degradation, and post-translational modification, and eventually evade the activation of cGAS-STING pathway.
Figure 2. Simplified diagram of the core element of the three unfolded protein response (UPR) signaling branches of the endoplasmic reticulum (ER). During different viral infections, the ER stress activates the three stress sensor proteins: IRE1α, ATF6, and PERK (Detailed in reviews [44, 64]). Each sensor uses a distinct mechanism of signal transduction to drive the transcription of UPR target genes and eventually work as feedback loops to mitigate the ER stress [44, 64]. Upon ER stresses, ATF6, a transcriptional factor, translocate into the Golgi compartment where it is cleaved by the site (1/2) protease. The N-terminal cytosolic domain of cleaved ATF6 is released into cytosol and then translocated into the nucleus where it binds to ER stress-response elements to activate target genes, including XBP-1 and C/EBP-homologous protein (CHOP) [43]. The activation of PERK inhibits general protein translation by the phosphorylation of eIF2α, enabling dedicated translation of transcripts, including ATF4, a key transducer. The IRE1 branch is regulated by non-conventional mRNA splicing [44, 64]. Subsequently, the activated IRE1 processes XBP1 mRNA to generate the spliced form of XBP1 protein (XBP1s), which participates in IRE1α-mediated UPR pathway in response to ER stresses [44, 64]. Eventually, the activation of the cleaved ATF6 (N-ATF6), ATF4, and XBP1 transcription factors increases the protein-folding capacity in the ER lumen. Meanwhile, IRE1 and PERK sensors also decrease the load of proteins entering the ER [44, 64].
Table 1. Viruses activate and exploit the UPR branch of ER for viral replication.
|
Viruses |
Family |
Genome Structure |
Virion Structure |
Viral Protein |
ATF6 |
PERK |
IRE1α |
Ref |
|
PRRSV |
Arteriviridae |
Linear, ssRNA(+) |
Enveloped; Spherical |
? |
× |
√ |
√ |
[55] |
|
IBV |
Coronaviridae |
Linear, ssRNA(+) |
Enveloped; Spherical |
? |
× |
× |
√ |
[65] |
|
DENV |
Flaviridae |
Linear, ssRNA(+) |
Enveloped; Spherical |
? |
× |
√ |
√ |
[66] |
|
JEV |
Flaviridae |
Linear, ssRNA(+) |
Enveloped; Spherical |
NS4B |
√ |
√ |
√ |
[67-69] |
|
TBEV |
Flaviridae |
Linear, ssRNA(+) |
Enveloped; Rounded |
? |
√ |
× |
√ |
[70] |
|
HCV |
Flaviridae |
Linear, ssRNA(+) |
Enveloped; Spherical |
? |
√ |
× |
× |
[71] |
|
IAV |
Orthomyxoviridae |
Segmented, ssRNA (-) |
Enveloped; Rounded |
? |
× |
|
√ |
[72] |
|
NDV |
Paramyxoviridae |
Linear, ssRNA(-) |
Enveloped; Spherical |
F and HN |
√ |
√ |
√ |
[56, 73] |
|
MCMV |
Herpesviridae |
Linear, ds DNA |
Enveloped; Spherical |
? |
? |
√ |
× |
[74, 75] |
|
HSV-1 |
Herpesviridae |
Linear, dsDNA |
Enveloped; Spherical |
UL41/ICP0/γ134.5 |
√ |
√ |
× |
[76-79] |
|
HBV |
Hepadnaviridae |
Circular dsDNA |
Enveloped; Spherical |
? |
√ |
× |
√ |
[80] |
During different viral infections, the ER stress activates the three stress sensor proteins: IRE1α, ATF6, and PERK (review in the references [44, 64]). The following abbreviations are used in this table: murine cytomegalovirus, MCMV; avian coronavirus infectious bronchitis virus, IBV; porcine reproductive and respiratory syndrome virus, PRRSV ; hepatitis C virus, HCV; hepatitis B virus, HBV; influenza A virus, IAV; human herpes simplex virus-1, HSV-1; dengue virus, DENV; tick-borne encephalitis virus, TBEV; Japanese encephalitis virus, JEV; Newcastle disease virus, NDV. The symbols √, ×, and ? indicate activation, inhibition, and unknown, respectively.
Rearrangement of peroxisome for infectious progeny
The peroxisomes are single membrane-bounded organelles that function in numerous metabolic pathways, including β-oxidation of long-chain fatty acids, detoxification of hydrogen peroxide, and synthesis of ether phospholipids and bile acids [81, 82]. Notably, the mitochondria and peroxisomes share common functions in the β-oxidation of fatty acids and the reduction of damaging peroxides. Proliferation of peroxisome is largely mediated by growth and division. Peroxisomal division in mammalian cells comprises multiple processes, including membrane deformation, elongation, constriction, and fission [83]. With the exception of peroxin (PEX)11, the peroxisomes and mitochondria share common fission machinery, including DRP-1, Mff, and Fis1 [84, 85]. The fission machinery of peroxisome is orchestrated by PEX-11β and mitochondrial fission factors [83]. Mitochondrial-derived vesicles (MDVs) are involved in the transportation of mitochondrial-anchored protein ligase (MAPL), a mitochondrial outer membrane, to peroxisomes [86]. The retromer complex containing vacuolar protein sorting (Vps) 5, Vps 26, and Vps 29, a known component of vesicle transport from the endosome to the Golgi apparatus, also regulates the transport of MAPL as a binding partner from the mitochondria to peroxisomes [87].
Viruses regulate the morphology and biogenesis of peroxisomes to promote progeny replication [1]. For instance, the C-terminal of the rotavirus VP4 protein is directly located in peroxisomes via its conserved peroxisomal targeting signal [88]. Meanwhile, viruses have exploited the myristoyl-CoA produced by peroxisome biogenesis for the N-myristoylation of viral proteins [1], such as ASFV [89], indicating that peroxisomal lipid metabolism contributes to viral replication. Another typical example is the tomato bushy stunt virus (TBSV), a member of the Tombusviridae family, which infects a variety of plant species. McCartney, A.W et al. reported that TBSV induced the rearrangement of peroxisomes and caused vesiculation of the peroxisomal membrane, where it provided a scaffold for virus anchoring and server as the sites of viral RNA synthesis [90]. In the absence of peroxisomes, TBSV also exploits the surface of the ER membrane as a viral factory for viral replication and assembly [91]. It is suggestive of the remarkable flexibility of the virus to use host membranes for replication.
Hijacking of Golgi apparatus for infectious progeny
The Golgi apparatus is a highly dynamic organelle whose function primarily includes saccule formation and utilization of such saccules in vesicle formation at the opposite face for delivery [92]. The normal cellular secretory pathway, bidirectional transport between the ER and Golgi apparatus, is mediated by tubulovesicular transport containers that depend on two coat protein complexes, COP-I and COP-II. The COP-II establishes a membrane flow from the ER to the Golgi complex [93]. However, the COP-I coat acts in retrograde transport from the Golgi back to the ER [94].
In general, certain viruses utilize the cellular trafficking and secretory pathway of the ER-Golgi transport system to replicate/release their progeny [3]. PV utilizes the components of the ADP-ribosylation factor (Arf) family of small GTPases [95] and cellular COP-II proteins [96] to form membrane-bound replication complex for viral replication, indicating that PV hijacks the components of the cellular secretory pathway for replication. As shown in Table 2, metonaviridae [23] and peribunyaviridae [24] hijack the Golgi complex to re-construct it as a viral factory for viral replication. For example, RUBV [23] and BUNV [24] infections modify cell structural morphology and remodel the Golgi complex as a viral factory during the entire life cycle. Furthermore, host secretory signaling is also crucial for innate and acquired immune responses, such as the exportation of proinflammatory and antiviral cytokines. Nearly 25 years ago, Doedens, J.R et al. reported that the 2B and 3A proteins of PV inhibited cellular protein secretion by directly blocking the transportation from the ER to the Golgi apparatus [97], indicating that the functional secretory protein is not indispensable for viral RNA replication. Mechanistically, Dodd, D.A et al. identified that the inhibition of 3A protein of PV on the ER to Golgi limited the antiviral cytokine secretion of native immune response and inflammation, including interleukin-6, interleukin-8, and β-interferon [98]. Deitz, S.B et al. also identified that PV 3A protein reduced the presentation of new antigens on the cell surface [99]. Considering that the ER adaptor STING was also located on the Golgi and ERGIC [61], we hypothesized that the membrane remodeling and modification of Golgi induced by viruses might also be involved in the regulation of cGAS-STING pathways (Figure 3). Collectively, all these data suggest that enteroviruses, such as PV and CVB, have evolved a series of strategies to hijack cellular trafficking and secretion for viral replication.
Table 2. Intracellular compartment sites for viral replication and assembly.
|
Family |
Viruses |
Genome Structure |
Virion Structure |
Replication Site |
Ref |
Assembly Site |
Ref |
||
|
Asfarviridae |
ASFV |
Liner, dsDNA |
Enveloped, spherical |
Nuclear and cytoplasmic |
[16, 100, 101] |
ER |
[54, 58] |
||
|
Poxviridae |
VV |
Liner, dsDNA |
Enveloped, brick-shaped |
cytoplasmic |
[102] |
ERGIC, ER |
[50, 51] |
||
|
Arteriviridae/Arteriviruses |
EAV, PRRSV |
Liner, ssRNA (+) |
Enveloped, spherical |
Cytoplasmic double membranes |
[52, 103] |
ER |
[52] |
||
|
Coronaviridae/Coronavirus |
SARS/MHV |
Liner, ssRNA (+) |
Enveloped, spherical |
Cytoplasmic double membranes |
[104] |
ERGIC, Golgi, ER |
[105, 106] |
||
|
Flaviviridae/ Hepacivirus |
HCV |
Linear, ssRNA(+) |
Enveloped, spherical |
Cytoplasmic |
[107] |
Autophagosome |
[108, 109] |
||
|
Metonaviridae/Rubivirus |
RUBV |
Liner, ssRNA (+) |
Enveloped, spherical isometric capsid? |
Cytoplasmic |
[23] |
Golgi, Lysosome |
[23, 110] |
||
|
Nodaviridae |
FHV |
Linear, ssRNA(+) |
Non-envelop, icosahedral |
Cytoplasmic |
[111, 112] |
OMM |
[111, 112] |
||
|
Togaviridae/Alphavirus |
SFV |
Liner, ssRNA (+) |
Enveloped, spherical and icosahedral |
Cytoplasmic |
[113, 114] |
Endosome /Lysosome |
[113-115] |
||
|
Tombusviridae |
TBSV |
Linear, ssRNA(+) |
Non-envelop, icosahedral |
Cytoplasmic |
[91] |
Peroxisome, ER |
[90, 91] |
||
|
Picornaviridae/Enterovirus |
CV/PV |
Linear, ssRNA(+) |
Non-envelop, spherical |
Cytoplasmic |
[116] |
ER |
[53, 116] |
||
|
Orthomyxoviridae |
IAV |
Segmented, ssRNA (-) |
Enveloped; Rounded |
Nuclear |
[117] |
ER, Golgi |
[117] |
||
|
Peribunyaviridae/Bunyavirus |
BUNV |
Segmented, ssRNA (-) |
Enveloped, spherical |
cytoplasmic |
[24] |
Golgi |
[24, 118] |
||
|
Retroviridae/Lentivirus |
HIV |
Linear, ssRNA(+) |
Enveloped, Spherical |
Nuclear |
[119] |
ER or Golgi |
[119] |
||
The following abbreviations are used in this table: African swine fever virus, ASFV; rubella virus, RUBV; Bunyamwera virus, BUNV; semliki forest virus, SFV; Flock house virus, FHV; tomato bushy stunt virus, TBSV; severe acute respiratory syndrome, SARS; murine hepatitis virus, MHV; porcine reproductive and respiratory syndrome virus, PRRSV ; equine arteritis virus, EAV; influenza A virus, IAV; human immune deficiency, HIV; vaccinia virus, VV; poliovirus, PV; coxsackieviruses, CV. reticulum-Golgi intermediate compartment, ERGIC; outer mitochondrial membranes, OMM.
Role of the lysosome and endosome in viral infections
The lysosome, an acidic and membrane-bound organelle, acts as a cellular recycling center and is filled with a number of hydrolases [120]. The lysosome-based degradation processes are subject to reciprocal regulation [121]. Lysosomes degrade unwanted materials which are delivered either from outside via the endocytic pathway or from inside via the autophagic pathway [121, 122]. For viral replication and assembly, certain viruses, including Alphaviruses [114], such as semliki forest virus (SFV) [113], exploit the membrane surface of the endosome and lysosome as a viral factory. Similarly, RUBV also can modify the membrane of lysosomes for a viral factory [110]. Meanwhile, the Toll-like receptors (TLR), such as TLR 3/7/9, are located on the endosome, indicating that the endosome also plays an important role in innate immunity [62]. Therefore, we speculate that another possible strategy is that viruses, particularly DNA viruses, evade the TLR-mediated activation of the NF-κB and transcription of proinflammatory cytokines. HBV infection suppresses TLR-9 expression and prevents TLR9 promoter activity in human primary B cells [123]. Interestingly, DENV, a positive-stand RNA virus, activates the TLR9 by inducing mtDNA release in human dendritic cells [124]. Additionally, the endosomal-lysosomal sorting system is a complex and dynamic vesicular sorting signaling, which is fundamental to maintain homeostasis [125]. Viruses, particularly enveloped viruses such as HIV [119], have evolved several strategies to hijack the endosomal sorting complex required for the transport (ESCRT) complex to facilitate viral budding. Collectively, all these data indicate that different viruses utilize different strategies to hijack the endosome/lysosome for viral progeny.
References
- Lazarow, P. B., Viruses exploiting peroxisomes. Current opinion in microbiology 2011, 14, (4), 458-69.
- Sanchez, E. L.; Lagunoff, M., Viral activation of cellular metabolism. Virology 2015, 479-480, 609-618.
- Robinson, M.; Schor, S.; Barouch-Bentov, R.; Einav, S., Viral journeys on the intracellular highways. Cellular and molecular life sciences : CMLS 2018.
- Twig, G.; Hyde, B.; Shirihai, O. S., Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochimica et biophysica acta 2008, 1777, (9), 1092-7.
- Chen, H.; Detmer, S. A.; Ewald, A. J.; Griffin, E. E.; Fraser, S. E.; Chan, D. C., Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. The Journal of cell biology 2003, 160, (2), 189-200.
- Song, Z.; Chen, H.; Fiket, M.; Alexander, C.; Chan, D. C., OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. The Journal of cell biology 2007, 178, (5), 749-55.
- Ehses, S.; Raschke, I.; Mancuso, G.; Bernacchia, A.; Geimer, S.; Tondera, D.; Martinou, J. C.; Westermann, B.; Rugarli, E. I.; Langer, T., Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. The Journal of cell biology 2009, 187, (7), 1023-36.
- Head, B.; Griparic, L.; Amiri, M.; Gandre-Babbe, S.; van der Bliek, A. M., Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. The Journal of cell biology 2009, 187, (7), 959-66.
- Olichon, A.; Baricault, L.; Gas, N.; Guillou, E.; Valette, A.; Belenguer, P.; Lenaers, G., Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. The Journal of biological chemistry 2003, 278, (10), 7743-6.
- Santel, A.; Fuller, M. T., Control of mitochondrial morphology by a human mitofusin. Journal of cell science 2001, 114, (Pt 5), 867-74.
- Cipolat, S.; Martins de Brito, O.; Dal Zilio, B.; Scorrano, L., OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proceedings of the National Academy of Sciences of the United States of America 2004, 101, (45), 15927-32.
- Manor, U.; Bartholomew, S.; Golani, G.; Christenson, E.; Kozlov, M.; Higgs, H.; Spudich, J.; Lippincott-Schwartz, J., A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. eLife 2015, 4.
- Smirnova, E.; Griparic, L.; Shurland, D. L.; van der Bliek, A. M., Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Molecular biology of the cell 2001, 12, (8), 2245-56.
- Lee, H.; Yoon, Y., Mitochondrial fission: regulation and ER connection. Molecules and cells 2014, 37, (2), 89-94.
- Otera, H.; Wang, C.; Cleland, M. M.; Setoguchi, K.; Yokota, S.; Youle, R. J.; Mihara, K., Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. The Journal of cell biology 2010, 191, (6), 1141-58.
- Castello, A.; Quintas, A.; Sanchez, E. G.; Sabina, P.; Nogal, M.; Carrasco, L.; Revilla, Y., Regulation of host translational machinery by African swine fever virus. PLoS pathogens 2009, 5, (8), e1000562.
- Wai, T.; Langer, T., Mitochondrial Dynamics and Metabolic Regulation. Trends in endocrinology and metabolism: TEM 2016, 27, (2), 105-17.
- Cogliati, S.; Frezza, C.; Soriano, M. E.; Varanita, T.; Quintana-Cabrera, R.; Corrado, M.; Cipolat, S.; Costa, V.; Casarin, A.; Gomes, L. C.; Perales-Clemente, E.; Salviati, L.; Fernandez-Silva, P.; Enriquez, J. A.; Scorrano, L., Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 2013, 155, (1), 160-71.
- Lebeau, J.; Saunders, J. M.; Moraes, V. W. R.; Madhavan, A.; Madrazo, N.; Anthony, M. C.; Wiseman, R. L., The PERK Arm of the Unfolded Protein Response Regulates Mitochondrial Morphology during Acute Endoplasmic Reticulum Stress. Cell reports 2018, 22, (11), 2827-2836.
- Chatel-Chaix, L.; Cortese, M.; Romero-Brey, I.; Bender, S.; Neufeldt, C. J.; Fischl, W.; Scaturro, P.; Schieber, N.; Schwab, Y.; Fischer, B.; Ruggieri, A.; Bartenschlager, R., Dengue Virus Perturbs Mitochondrial Morphodynamics to Dampen Innate Immune Responses. Cell host & microbe 2016, 20, (3), 342-356.
- Shi, C. S.; Qi, H. Y.; Boularan, C.; Huang, N. N.; Abu-Asab, M.; Shelhamer, J. H.; Kehrl, J. H., SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. Journal of immunology (Baltimore, Md. : 1950) 2014, 193, (6), 3080-9.
- Fontaine, K. A.; Camarda, R.; Lagunoff, M., Vaccinia virus requires glutamine but not glucose for efficient replication. Journal of virology 2014, 88, (8), 4366-74.
- Fontana, J.; Lopez-Iglesias, C.; Tzeng, W. P.; Frey, T. K.; Fernandez, J. J.; Risco, C., Three-dimensional structure of Rubella virus factories. Virology 2010, 405, (2), 579-91.
- Fontana, J.; Lopez-Montero, N.; Elliott, R. M.; Fernandez, J. J.; Risco, C., The unique architecture of Bunyamwera virus factories around the Golgi complex. Cellular microbiology 2008, 10, (10), 2012-28.
- Rainbolt, T. K.; Saunders, J. M.; Wiseman, R. L., Stress-responsive regulation of mitochondria through the ER unfolded protein response. Trends in endocrinology and metabolism: TEM 2014, 25, (10), 528-37.
- Chan, D. C., Fusion and fission: interlinked processes critical for mitochondrial health. Annual review of genetics 2012, 46, 265-87.
- Castanier, C.; Garcin, D.; Vazquez, A.; Arnoult, D., Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO reports 2010, 11, (2), 133-8.
- Yasukawa, K.; Oshiumi, H.; Takeda, M.; Ishihara, N.; Yanagi, Y.; Seya, T.; Kawabata, S.; Koshiba, T., Mitofusin 2 inhibits mitochondrial antiviral signaling. Science signaling 2009, 2, (84), ra47.
- Onoguchi, K.; Onomoto, K.; Takamatsu, S.; Jogi, M.; Takemura, A.; Morimoto, S.; Julkunen, I.; Namiki, H.; Yoneyama, M.; Fujita, T., Virus-infection or 5'ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS pathogens 2010, 6, (7), e1001012.
- Kawai, T.; Akira, S., Innate immune recognition of viral infection. Nature immunology 2006, 7, (2), 131-7.
- Martinvalet, D., The role of the mitochondria and the endoplasmic reticulum contact sites in the development of the immune responses. Cell death & disease 2018, 9, (3), 336.
- Yoshizumi, T.; Ichinohe, T.; Sasaki, O.; Otera, H.; Kawabata, S.; Mihara, K.; Koshiba, T., Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nature communications 2014, 5, 4713.
- Xia, M.; Gonzalez, P.; Li, C.; Meng, G.; Jiang, A.; Wang, H.; Gao, Q.; Debatin, K. M.; Beltinger, C.; Wei, J., Mitophagy enhances oncolytic measles virus replication by mitigating DDX58/RIG-I-like receptor signaling. Journal of virology 2014, 88, (9), 5152-64.
- Kim, S. J.; Syed, G. H.; Khan, M.; Chiu, W. W.; Sohail, M. A.; Gish, R. G.; Siddiqui, A., Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proceedings of the National Academy of Sciences of the United States of America 2014, 111, (17), 6413-8.
- Kim, S. J.; Syed, G. H.; Siddiqui, A., Hepatitis C virus induces the mitochondrial translocation of Parkin and subsequent mitophagy. PLoS pathogens 2013, 9, (3), e1003285.
- Kim, S. J.; Khan, M.; Quan, J.; Till, A.; Subramani, S.; Siddiqui, A., Hepatitis B virus disrupts mitochondrial dynamics: induces fission and mitophagy to attenuate apoptosis. PLoS pathogens 2013, 9, (12), e1003722.
- Dreux, M.; Gastaminza, P.; Wieland, S. F.; Chisari, F. V., The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A 2009, 106, (33), 14046-51.
- Romero-Brey, I.; Bartenschlager, R., Endoplasmic Reticulum: The Favorite Intracellular Niche for Viral Replication and Assembly. Viruses 2016, 8, (6).
- Powers, R. E.; Wang, S.; Liu, T. Y.; Rapoport, T. A., Reconstitution of the tubular endoplasmic reticulum network with purified components. Nature 2017, 543, (7644), 257-260.
- Verchot, J., How does the stressed out ER find relief during virus infection? Current opinion in virology 2016, 17, 74-79.
- Wang, M.; Kaufman, R. J., Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 2016, 529, (7586), 326-35.
- Ruggiano, A.; Foresti, O.; Carvalho, P., Quality control: ER-associated degradation: protein quality control and beyond. The Journal of cell biology 2014, 204, (6), 869-79.
- Guo, F. J.; Xiong, Z.; Lu, X.; Ye, M.; Han, X.; Jiang, R., ATF6 upregulates XBP1S and inhibits ER stress-mediated apoptosis in osteoarthritis cartilage. Cellular signalling 2014, 26, (2), 332-42.
- Walter, P.; Ron, D., The unfolded protein response: from stress pathway to homeostatic regulation. Science (New York, N.Y.) 2011, 334, (6059), 1081-6.
- Fujita, E.; Kouroku, Y.; Isoai, A.; Kumagai, H.; Misutani, A.; Matsuda, C.; Hayashi, Y. K.; Momoi, T., Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II). Human molecular genetics 2007, 16, (6), 618-29.
- Moon, S. L.; Barnhart, M. D.; Wilusz, J., Inhibition and avoidance of mRNA degradation by RNA viruses. Current opinion in microbiology 2012, 15, (4), 500-505.
- Hsu, N. Y.; Ilnytska, O.; Belov, G.; Santiana, M.; Chen, Y. H.; Takvorian, P. M.; Pau, C.; van der Schaar, H.; Kaushik-Basu, N.; Balla, T.; Cameron, C. E.; Ehrenfeld, E.; van Kuppeveld, F. J.; Altan-Bonnet, N., Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 2010, 141, (5), 799-811.
- Tenorio, R.; Fernandez de Castro, I.; Knowlton, J. J.; Zamora, P. F.; Lee, C. H.; Mainou, B. A.; Dermody, T. S.; Risco, C., Reovirus sigmaNS and muNS Proteins Remodel the Endoplasmic Reticulum to Build Replication Neo-Organelles. mBio 2018, 9, (4).
- Miller, S.; Kastner, S.; Krijnse-Locker, J.; Buhler, S.; Bartenschlager, R., The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. The Journal of biological chemistry 2007, 282, (12), 8873-82.
- Risco, C.; Rodriguez, J. R.; Lopez-Iglesias, C.; Carrascosa, J. L.; Esteban, M.; Rodriguez, D., Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. Journal of virology 2002, 76, (4), 1839-55.
- Husain, M.; Moss, B., Evidence against an essential role of COPII-mediated cargo transport to the endoplasmic reticulum-Golgi intermediate compartment in the formation of the primary membrane of vaccinia virus. Journal of virology 2003, 77, (21), 11754-66.
- Knoops, K.; Barcena, M.; Limpens, R. W.; Koster, A. J.; Mommaas, A. M.; Snijder, E. J., Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. Journal of virology 2012, 86, (5), 2474-87.
- Suhy, D. A.; Giddings, T. H., Jr.; Kirkegaard, K., Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. Journal of virology 2000, 74, (19), 8953-65.
- Rodriguez, J. M.; Garcia-Escudero, R.; Salas, M. L.; Andres, G., African swine fever virus structural protein p54 is essential for the recruitment of envelope precursors to assembly sites. Journal of virology 2004, 78, (8), 4299-1313.
- Gao, P.; Chai, Y.; Song, J.; Liu, T.; Chen, P.; Zhou, L.; Ge, X.; Guo, X.; Han, J.; Yang, H., Reprogramming the unfolded protein response for replication by porcine reproductive and respiratory syndrome virus. PLoS pathogens 2019, 15, (11), e1008169.
- Ren, S.; Rehman, Z. U.; Shi, M.; Yang, B.; Liu, P.; Yin, Y.; Qu, Y.; Meng, C.; Yang, Z.; Gao, X.; Sun, Y.; Ding, C., Hemagglutinin-neuraminidase and fusion proteins of virulent Newcastle disease virus cooperatively disturb fusion–fission homeostasis to enhance mitochondrial function by activating the unfolded protein response of endoplasmic reticulum and mitochondrial stress. Veterinary research 2019, 50, (1).
- Targett-Adams, P.; Boulant, S.; Douglas, M. W.; McLauchlan, J., Lipid metabolism and HCV infection. Viruses 2010, 2, (5), 1195-217.
- Cobbold, C.; Brookes, S. M.; Wileman, T., Biochemical requirements of virus wrapping by the endoplasmic reticulum: involvement of ATP and endoplasmic reticulum calcium store during envelopment of African swine fever virus. Journal of virology 2000, 74, (5), 2151-60.
- Liu, X.; Palaniyandi, S.; Zhu, I.; Tang, J.; Li, W.; Wu, X.; Ochsner, S. P.; Pauza, C. D.; Cohen, J. I.; Zhu, X., Human cytomegalovirus evades antibody-mediated immunity through endoplasmic reticulum-associated degradation of the FcRn receptor. Nature communications 2019, 10, (1), 3020.
- Jung, K. I.; Ko, D. H.; Shin, N.; Pyo, C. W.; Choi, S. Y., Endoplasmic reticulum-associated degradation potentiates the infectivity of influenza A virus by regulating the host redox state. Free radical biology & medicine 2019, 135, 293-305.
- Chen, Q.; Sun, L.; Chen, Z. J., Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nature immunology 2016, 17, (10), 1142-9.
- Motwani, M.; Pesiridis, S.; Fitzgerald, K. A., DNA sensing by the cGAS-STING pathway in health and disease. Nat Rev Genet 2019, 20, (11), 657-674.
- Ishikawa, H.; Barber, G. N., STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008, 455, (7213), 674-8.
- Ron, D.; Walter, P., Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007, 8, (7), 519-29.
- Fung, T. S.; Liao, Y.; Liu, D. X., The endoplasmic reticulum stress sensor IRE1alpha protects cells from apoptosis induced by the coronavirus infectious bronchitis virus. Journal of virology 2014, 88, (21), 12752-64.
- Lee, Y. R.; Kuo, S. H.; Lin, C. Y.; Fu, P. J.; Lin, Y. S.; Yeh, T. M.; Liu, H. S., Dengue virus-induced ER stress is required for autophagy activation, viral replication, and pathogenesis both in vitro and in vivo. Scientific reports 2018, 8, (1), 489.
- Wang, Q.; Xin, X.; Wang, T.; Wan, J.; Ou, Y.; Yang, Z.; Yu, Q.; Zhu, L.; Guo, Y.; Wu, Y.; Ding, Z.; Zhang, Y.; Pan, Z.; Tang, Y.; Li, S.; Kong, L., Japanese Encephalitis Virus Induces Apoptosis and Encephalitis by Activating the PERK Pathway. Journal of virology 2019, 93, (17).
- Huang, M.; Xu, A.; Wu, X.; Zhang, Y.; Guo, Y.; Guo, F.; Pan, Z.; Kong, L., Japanese encephalitis virus induces apoptosis by the IRE1/JNK pathway of ER stress response in BHK-21 cells. Arch Virol 2016, 161, (3), 699-703.
- Sharma, M.; Bhattacharyya, S.; Sharma, K. B.; Chauhan, S.; Asthana, S.; Abdin, M. Z.; Vrati, S.; Kalia, M., Japanese encephalitis virus activates autophagy through XBP1 and ATF6 ER stress sensors in neuronal cells. The Journal of general virology 2017, 98, (5), 1027-1039.
- Yu, C.; Achazi, K.; Niedrig, M., Tick-borne encephalitis virus triggers inositol-requiring enzyme 1 (IRE1) and transcription factor 6 (ATF6) pathways of unfolded protein response. Virus research 2013, 178, (2), 471-7.
- Tardif, K. D.; Mori, K.; Kaufman, R. J.; Siddiqui, A., Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. The Journal of biological chemistry 2004, 279, (17), 17158-64.
- Hassan, I. H.; Zhang, M. S.; Powers, L. S.; Shao, J. Q.; Baltrusaitis, J.; Rutkowski, D. T.; Legge, K.; Monick, M. M., Influenza A viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IRE1) stress pathway. The Journal of biological chemistry 2012, 287, (7), 4679-89.
- Li, Y.; Jiang, W.; Niu, Q.; Sun, Y.; Meng, C.; Tan, L.; Song, C.; Qiu, X.; Liao, Y.; Ding, C., eIF2alpha-CHOP-BCl-2/JNK and IRE1alpha-XBP1/JNK signaling promote apoptosis and inflammation and support the proliferation of Newcastle disease virus. Cell death & disease 2019, 10, (12), 891.
- Stahl, S.; Burkhart, J. M.; Hinte, F.; Tirosh, B.; Mohr, H.; Zahedi, R. P.; Sickmann, A.; Ruzsics, Z.; Budt, M.; Brune, W., Cytomegalovirus downregulates IRE1 to repress the unfolded protein response. PLoS pathogens 2013, 9, (8), e1003544.
- Qian, Z.; Xuan, B.; Chapa, T. J.; Gualberto, N.; Yu, D., Murine cytomegalovirus targets transcription factor ATF4 to exploit the unfolded-protein response. Journal of virology 2012, 86, (12), 6712-23.
- Burnett, H. F.; Audas, T. E.; Liang, G.; Lu, R. R., Herpes simplex virus-1 disarms the unfolded protein response in the early stages of infection. Cell stress & chaperones 2012, 17, (4), 473-83.
- Zhang, P.; Su, C.; Jiang, Z.; Zheng, C., Herpes Simplex Virus 1 UL41 Protein Suppresses the IRE1/XBP1 Signal Pathway of the Unfolded Protein Response via Its RNase Activity. Journal of virology 2017, 91, (4).
- Cheng, G.; Feng, Z.; He, B., Herpes simplex virus 1 infection activates the endoplasmic reticulum resident kinase PERK and mediates eIF-2alpha dephosphorylation by the gamma(1)34.5 protein. Journal of virology 2005, 79, (3), 1379-88.
- Mulvey, M.; Arias, C.; Mohr, I., Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. Journal of virology 2007, 81, (7), 3377-90.
- Li, B.; Gao, B.; Ye, L.; Han, X.; Wang, W.; Kong, L.; Fang, X.; Zeng, Y.; Zheng, H.; Li, S.; Wu, Z.; Ye, L., Hepatitis B virus X protein (HBx) activates ATF6 and IRE1-XBP1 pathways of unfolded protein response. Virus research 2007, 124, (1-2), 44-9.
- Wanders, R. J.; Waterham, H. R., Biochemistry of mammalian peroxisomes revisited. Annual review of biochemistry 2006, 75, 295-332.
- Till, A.; Lakhani, R.; Burnett, S. F.; Subramani, S., Pexophagy: the selective degradation of peroxisomes. International journal of cell biology 2012, 2012, 512721.
- Honsho, M.; Yamashita, S.-i.; Fujiki, Y., Peroxisome homeostasis: Mechanisms of division and selective degradation of peroxisomes in mammals. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2016, 1863, (5), 984-991.
- Delille, H. K.; Schrader, M., Targeting of hFis1 to peroxisomes is mediated by Pex19p. The Journal of biological chemistry 2008, 283, (45), 31107-15.
- Schrader, M., Shared components of mitochondrial and peroxisomal division. Biochimica et biophysica acta 2006, 1763, (5-6), 531-41.
- Neuspiel, M.; Schauss, A. C.; Braschi, E.; Zunino, R.; Rippstein, P.; Rachubinski, R. A.; Andrade-Navarro, M. A.; McBride, H. M., Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Current biology : CB 2008, 18, (2), 102-8.
- Braschi, E.; Goyon, V.; Zunino, R.; Mohanty, A.; Xu, L.; McBride, H. M., Vps35 mediates vesicle transport between the mitochondria and peroxisomes. Current biology : CB 2010, 20, (14), 1310-5.
- Mohan, K. V.; Som, I.; Atreya, C. D., Identification of a type 1 peroxisomal targeting signal in a viral protein and demonstration of its targeting to the organelle. Journal of virology 2002, 76, (5), 2543-7.
- Aguado, B.; Vinuela, E.; Alcami, A., African swine fever virus fatty acid acylated proteins. Virology 1991, 185, (2), 942-5.
- McCartney, A. W.; Greenwood, J. S.; Fabian, M. R.; White, K. A.; Mullen, R. T., Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. The Plant cell 2005, 17, (12), 3513-31.
- Jonczyk, M.; Pathak, K. B.; Sharma, M.; Nagy, P. D., Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology 2007, 362, (2), 320-30.
- James Morre, D.; Mollenhauer, H. H., Microscopic morphology and the origins of the membrane maturation model of Golgi apparatus function. Int Rev Cytol 2007, 262, 191-218.
- Barlowe, C.; Orci, L.; Yeung, T.; Hosobuchi, M.; Hamamoto, S.; Salama, N.; Rexach, M. F.; Ravazzola, M.; Amherdt, M.; Schekman, R., COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 1994, 77, (6), 895-907.
- Rabouille, C.; Klumperman, J., Opinion: The maturing role of COPI vesicles in intra-Golgi transport. Nature reviews. Molecular cell biology 2005, 6, (10), 812-7.
- Belov, G. A.; Altan-Bonnet, N.; Kovtunovych, G.; Jackson, C. L.; Lippincott-Schwartz, J.; Ehrenfeld, E., Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. Journal of virology 2007, 81, (2), 558-67.
- Rust, R. C.; Landmann, L.; Gosert, R.; Tang, B. L.; Hong, W.; Hauri, H. P.; Egger, D.; Bienz, K., Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. Journal of virology 2001, 75, (20), 9808-18.
- Doedens, J. R.; Kirkegaard, K., Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. The EMBO journal 1995, 14, (5), 894-907.
- Dodd, D. A.; Giddings, T. H., Jr.; Kirkegaard, K., Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. Journal of virology 2001, 75, (17), 8158-65.
- Deitz, S. B.; Dodd, D. A.; Cooper, S.; Parham, P.; Kirkegaard, K., MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proceedings of the National Academy of Sciences of the United States of America 2000, 97, (25), 13790-5.
- Garcia-Beato, R.; Salas, M. L.; Vinuela, E.; Salas, J., Role of the host cell nucleus in the replication of African swine fever virus DNA. Virology 1992, 188, (2), 637-49.
- Dixon, L. K.; Chapman, D. A.; Netherton, C. L.; Upton, C., African swine fever virus replication and genomics. Virus research 2013, 173, (1), 3-14.
- Katsafanas, G. C.; Moss, B., Colocalization of Transcription and Translation within Cytoplasmic Poxvirus Factories Coordinates Viral Expression and Subjugates Host Functions. Cell host & microbe 2007, 2, (4), 221-228.
- Pedersen, K. W.; van der Meer, Y.; Roos, N.; Snijder, E. J., Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J Virol 1999, 73, (3), 2016-26.
- Angelini, M. M.; Akhlaghpour, M.; Neuman, B. W.; Buchmeier, M. J.; Moscona, A., Severe Acute Respiratory Syndrome Coronavirus Nonstructural Proteins 3, 4, and 6 Induce Double-Membrane Vesicles. mBio 2013, 4, (4).
- Bost, A. G.; Prentice, E.; Denison, M. R., Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology 2001, 285, (1), 21-9.
- Stertz, S.; Reichelt, M.; Spiegel, M.; Kuri, T.; Martinez-Sobrido, L.; Garcia-Sastre, A.; Weber, F.; Kochs, G., The intracellular sites of early replication and budding of SARS-coronavirus. Virology 2007, 361, (2), 304-15.
- Wang, L.; Tian, Y.; Ou, J. H., HCV induces the expression of Rubicon and UVRAG to temporally regulate the maturation of autophagosomes and viral replication. PLoS pathogens 2015, 11, (3), e1004764.
- Sir, D.; Kuo, C. F.; Tian, Y.; Liu, H. M.; Huang, E. J.; Jung, J. U.; Machida, K.; Ou, J. H., Replication of hepatitis C virus RNA on autophagosomal membranes. The Journal of biological chemistry 2012, 287, (22), 18036-43.
- Mohl, B. P.; Bartlett, C.; Mankouri, J.; Harris, M., Early events in the generation of autophagosomes are required for the formation of membrane structures involved in hepatitis C virus genome replication. The Journal of general virology 2016, 97, (3), 680-693.
- Magliano, D.; Marshall, J. A.; Bowden, D. S.; Vardaxis, N.; Meanger, J.; Lee, J. Y., Rubella virus replication complexes are virus-modified lysosomes. Virology 1998, 240, (1), 57-63.
- Miller, D. J.; Schwartz, M. D.; Ahlquist, P., Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. Journal of virology 2001, 75, (23), 11664-76.
- Ertel, K. J.; Benefield, D.; Castano-Diez, D.; Pennington, J. G.; Horswill, M.; den Boon, J. A.; Otegui, M. S.; Ahlquist, P., Cryo-electron tomography reveals novel features of a viral RNA replication compartment. eLife 2017, 6.
- Kujala, P.; Ikaheimonen, A.; Ehsani, N.; Vihinen, H.; Auvinen, P.; Kaariainen, L., Biogenesis of the Semliki Forest Virus RNA Replication Complex. Journal of virology 2001, 75, (8), 3873-3884.
- Froshauer, S.; Kartenbeck, J.; Helenius, A., Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. The Journal of cell biology 1988, 107, (6 Pt 1), 2075-86.
- Spuul, P.; Balistreri, G.; Kaariainen, L.; Ahola, T., Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest Virus replication complexes from the plasma membrane to modified lysosomes. Journal of virology 2010, 84, (15), 7543-57.
- Baggen, J.; Thibaut, H. J.; Strating, J.; van Kuppeveld, F. J. M., The life cycle of non-polio enteroviruses and how to target it. Nature reviews. Microbiology 2018, 16, (6), 368-381.
- Long, J. S.; Mistry, B.; Haslam, S. M.; Barclay, W. S., Host and viral determinants of influenza A virus species specificity. Nature reviews. Microbiology 2019, 17, (2), 67-81.
- Shi, X.; Kohl, A.; Leonard, V. H.; Li, P.; McLees, A.; Elliott, R. M., Requirement of the N-terminal region of orthobunyavirus nonstructural protein NSm for virus assembly and morphogenesis. Journal of virology 2006, 80, (16), 8089-99.
- Gomez, C.; Hope, T. J., The ins and outs of HIV replication. Cellular microbiology 2005, 7, (5), 621-6.
- Aits, S.; Jaattela, M., Lysosomal cell death at a glance. Journal of cell science 2013, 126, (Pt 9), 1905-12.
- Perera, R. M.; Zoncu, R., The Lysosome as a Regulatory Hub. Annual review of cell and developmental biology 2016, 32, 223-253.
- Saftig, P.; Klumperman, J., Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nature reviews. Molecular cell biology 2009, 10, (9), 623-35.
- Tout, I.; Gomes, M.; Ainouze, M.; Marotel, M.; Pecoul, T.; Durantel, D.; Vaccarella, S.; Dubois, B.; Loustaud-Ratti, V.; Walzer, T.; Alain, S.; Chemin, I.; Hasan, U., Hepatitis B Virus Blocks the CRE/CREB Complex and Prevents TLR9 Transcription and Function in Human B Cells. Journal of immunology (Baltimore, Md. : 1950) 2018, 201, (8), 2331-2344.
- Lai, J. H.; Wang, M. Y.; Huang, C. Y.; Wu, C. H.; Hung, L. F.; Yang, C. Y.; Ke, P. Y.; Luo, S. F.; Liu, S. J.; Ho, L. J., Infection with the dengue RNA virus activates TLR9 signaling in human dendritic cells. EMBO reports 2018, 19, (8).
- Bonifacino, J. S.; Traub, L. M., Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annual review of biochemistry 2003, 72, 395-447.
|
© 2020 by the authors. Submitted for possible open access publication under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/). |
This entry is adapted from the peer-reviewed paper 10.3390/ijms21103689