Peroxiredoxin (Prx) is a relatively recently discovered antioxidant enzyme family that scavenges peroxides and is known to be present in organisms from biological taxa ranging from bacteria to multicellular eukaryotes, including photosynthetic organisms.
- 2-Cys peroxiredoxin
- antioxidant enzyme
- growth potential
- photosynthesis
1. Introduction
Peroxiredoxin (Prx) is a recently discovered antioxidant enzyme family [1] involved in various signaling pathways as well as antioxidant activity based on peroxide-scavenging ability [2] and is known to be present in organisms found in biological taxa ranging from bacteria to multicellular eukaryotes [3][4][5]. Generally, Prx enzymes can be classified into several groups based on the amino-acid sequence and locations of the cysteine residues. Plants and cyanobacteria have several types of Prx enzymes: 2-CysPrx, 1-CysPrx, PrxQ, and PrxII (or type II Prx), all of which are targeted to different organelles [4]. For example, 2-CysPrx and PrxQ are located in the chloroplast; 1-CysPrx is located in the nucleus and cytoplasm; and PrxIIs are located in the cytosol, mitochondria, and plastids. The Prx enzymes located in the chloroplast of photosynthetic organisms (2-CysPrx and PrxQ) contribute to the removal of hazardous reactive oxygen species (ROS) produced through photosynthetic processes, and they are involved in redox signal control through mechanisms such as retrograde signaling [6]. Many Prx studies have, therefore, involved land plants; cyanobacteria [4]; and eukaryotic, unicellular algae, especially green algae [7]. These studies have gradually revealed the role of Prx enzymes in protecting organisms from various environmental stresses [8] and controlling redox signals [9].
In contrast, there have been few studies of antioxidant systems, including those that involve Prx, in the eukaryotic algae that cause harmful algal blooms (HABs)—mainly raphidophytes and dinoflagellates—and induce the mass mortality of aquacultured fish. The raphidophyte genus Chattonella (Figure 1), in particular, has caused huge economic losses to the aquaculture industry in Japan: approximately JPY 7.1 billion in 1972 in the Harima-nada marine region [10], for example, as well as JPY 2.9 billion in 2009 and JPY 5.3 billion in 2010 in the Yatsushiro Sea [11]. This genus has also caused the mass mortality of cultured southern bluefin tuna in Australia [12] and of salmon in Norway [13]. In the field of HAB studies, it is essential to clarify the environmental factors (e.g., temperature, light intensity, and nutrient concentrations) that affect algal growth and physiology in order to predict bloom dynamics and thereby mitigate damage to fisheries.

Figure 1. An image of Chattonella marina var. antiqua.
Many laboratory [14][15] and field [16][17] studies have investigated the effects of environmental factors on the growth of Chattonella. Generally, the formation of phytoplankton blooms is affected by factors such as algal growth, predation pressure, and physical transport processes [18][19][20]. C. marina has been reported to grow under a broad range of temperature (15–30 °C) and salinity (10–35) conditions, and its growth rate has been reported to saturate at light intensities of 110 µmol photons m–2 s–1 [20]. Qiu et al. [21] reported that C. marina is capable of growing under severe environmental conditions, including an irradiance as high as ~1000 µmol m–2 s–1, which is comparable to the average irradiance in surface seawater on a sunny, mid-summer day in coastal areas. However, excessive irradiance is harmful to photosynthetic organisms and may cause high rates of production of ROS. The implication is that the physiology of C. marina enables it to resist light-induced oxidative stress during growth and thereby allows it to frequently form dense midsummer blooms in the coastal marine waters of Western Japan. Although Portune et al. [22] investigated antioxidant activity of harmful raphidophytes, few molecular biological studies related to antioxidant activities in HAB species, including C. marina, have been undertaken until recently.
A link between antioxidant enzymes and stress tolerance has been recognized for a broad range of organisms, including photosynthetic species. Miao et al. [23] reported that a mutant of the thale cress, Arabidopsis thaliana, contains a glutathione peroxidase (GPX) that enables it to tolerate a higher rate of water loss when stressed under drought conditions; this strain exhibits higher sensitivity to hydrogen peroxide (H2O2) treatment during seed germination and seedling development and produces more H2O2 in guard cells than wild-type strains. Insertion into the yeast of genes from the microalga Chlorella vulgaris Beijerinck that encode the antioxidant enzyme peroxiredoxin and NADPH-dependent thioredoxin reductase increases the tolerance of the yeast to freezing, heat, and oxidative stresses [24]. Knock-down strains of rice that contain functional ascorbate peroxidase (APX) isoforms exhibit earlier senescence than wild-type strains [25]. Antioxidant enzymes may thus be closely related to abiotic stress tolerance and growth potential.
C. marina generates higher amounts of ROS, such as the superoxide anion (O2−) and H2O2, than other unicellular algae [22][26]. Growth activity in this species is also inhibited by decomposition of O2− and H2O2 caused by addition of superoxide dismutase (SOD) and catalase (CAT) [27]. A certain level of oxidative stress by O2− and H2O2 might, therefore, have a beneficial effect on C. marina growth. However, because ROS attack various cellular molecules such as DNA, RNA, proteins, and lipids [28][29], high concentrations of ROS can have deleterious effects on growth. In mammals and plants, lipid peroxidation is caused by oxidative stress from ROS and non-radical reactions mediated by lipoxygenase [30][31][32]. Therefore, C. marina, which generates relatively high amounts of ROS compared to other unicellular algae, is probably exposed to a high degree of oxidative stress, and it might protect itself from ROS with a sophisticated system involving the use of antioxidants.
In many cases, HAB species form dense blooms during the summer under harsh environmental conditions characterized by supra-optimal irradiance and temperature. Understanding the physiological mechanisms that allow algae to tolerate these environmental conditions is important to biological and fisheries science. Recently, omics analysis has begun to be used to investigate the comprehensive molecular mechanisms that account for HAB phenomena [21][33][34][35][36][37][38]. Our studies have focused on the molecular mechanisms that control the growth of HAB species. Through our proteomic study, we identified proteins with a growth rate-dependent expression pattern, and we found that 2-CysPrx [21] was a possible key factor that enabled the HAB species C. marina to maintain a high cell division rate.
2. Temporal Changes of Levels of 2-CysPRX Enzymes During Algal Growth
In a previous proteomic study using two-dimensional electrophoresis [21], we detected a highly expressed protein in C. marina var. antiqua (NIES-1 strain) that was identified as 2-CysPrx based on the 20 residues of its N-terminal sequence (Figure 2). The abundance of the C. marina 2-CysPrx enzyme was highest during exponential growth when photosynthetic activity was high; the abundance then decreased roughly twofold during the late stationary phase (Figure 2). This protein accounted for 4% of all the proteins detected in an exponential-growth-phase sample of C. marina cells based on the fluorescence intensity of all protein spots in the 2-DE profile. If consideration is given to the difference between measurement methods, our estimation of the protein abundance was nearly equal to or higher than estimates reported in previous studies, which have indicated that the contributions of Prx (0.5–1%) to cellular protein are relatively high in several types of cultured cells [39]. The 2-CysPrx in C. marina is, therefore, highly expressed at the protein level and is thought to facilitate cell proliferation by removing peroxides produced during photosynthesis.
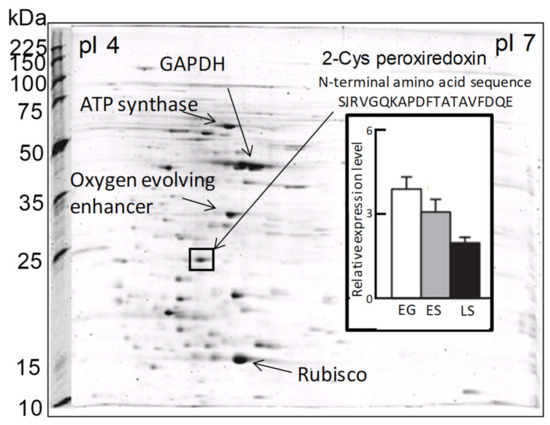
Figure 2. Two-dimensional gel electrophoresis profiles of Chattonella marina var. antiqua and some identified major protein spots, including 2-CysPrx. The bar graph shows the levels of 2-CysPrx protein expression in the exponential growth phase (EG), early stationary phase (ES), and late stationary phase (LS) [21].
A growth phase-dependent change of the level of expression of the 2-CysPrx protein and photosynthetic activity has also been observed during a natural bloom [40][41]. We investigated temporal variations of protein expression profiles in C. marina cells during a HAB (5–14 September 2012) that occurred in the inner part of the Ariake Sea to the north of the island of Kyushu, Japan (Figure 3A,B). Proteomic analyses revealed a decreasing trend in the abundances of 2-CysPRX (Figure 3C) and other photosynthesis-related proteins such as LHCP 4, Cyt c553, and GAPDH [41] as the bloom progressed. The maximum potential quantum yield of photosystem II (Fv/Fm ratio) decreased in a manner similar to the daily growth rate, which was calculated from the change of cell concentrations in a growth chamber between the sampling day and the next day (Figure 3D,E). The abundances of the above proteins were significantly (p < 0.05) and positively correlated with Fv/Fm ratios and daily growth rates [41].

Figure 3. Field sites for sampling from 5 to 18 September 2012 in the inner part of the Ariake Sea, Japan (A). Stars indicate the sampling stations. Temporal variations of the (B) cell concentrations, (C) relative 2-CysPrx expression levels, (D) photosynthetic activities (Fv/Fm ratios), and (E) growth of Chattonella marina cells in natural seawater samples. Growth rates were calculated from the change of cell concentrations between the sampling day and the next day after incubation in a growth chamber. Symbols indicate values for each station, and lines indicate the average values of all stations on the same sampling day [40][41].
The antioxidant function of 2-CysPrx in photosynthetic organisms has been investigated in higher plants [42][43][44][45] and cyanobacteria [3][46][47]. Expression of the 2-CysPrx genes is under developmental control, and steady-state mRNA levels decrease with tissue age in barley [43], Arabidopsis thaliana [44], and Riccia fluitans [45]. In cyanobacteria, the Fv/Fm ratio and growth rate under high irradiance are lower in a mutant strain of Synechocystis 6803 lacking 2-CysPrx than in wild-type cells [46]. Our discovery that the abundance of 2-CysPrx was positively correlated with both Fv/Fm ratios and growth rates was consistent with the results of those earlier studies. Chattonella is known to produce high levels of ROS as a byproduct of photosynthesis [22][48] or possibly of another pathway [49], and they, therefore, require large amounts of antioxidant enzymes to prevent damage from ROS during active growth. As a Chattonella bloom progresses, relatively low amounts of 2-CysPrx may provide insufficient protection, and the resultant accumulation of peroxides within cells can impair photosynthesis by damaging chloroplast structures [44] and cause a rapid termination of the bloom (Figure 3B) by triggering peroxide signaling events that lead to cell death [50]. Thus, further discussions are needed concerning the growth phase-dependent physiological function of 2-CysPrx content.
3. Structure of the 2-CysPrx Gene in Chattonella marina
To elucidate the mechanism responsible for the production of 2-CysPrx, we determined the structure of the 2-CysPrx gene and the predicted amino acid sequence using the rapid amplification of cDNA ends (RACE) method [51]. The open reading frame of the 2-CysPrx gene was 585 base pairs (bp) long and encoded a protein consisting of 195 amino acids. The putative amino acid sequence contained two cysteine residues located at the 49th and 170th amino acid positions from the N-terminal methionine residue. The sequence also possessed 2-CysPrx characteristic motifs, F (FFYPLDFTFVCPTEI), GGLG, EVCP, and YF [50]. We concluded that the 2-CysPrx gene is located in the chloroplast genome, because there was no intron in the DNA sequence, and the position of the 2-CysPrx gene relative to several other genes (ycf59–2-CysPrx–rpl35–rpl20) was the same as the position of the corresponding gene in the chloroplast genome of the raphidophyte Heterosigma akashiwo (GenBank accession no. EU168191.1) [52]. In addition, inverse polymerase chain reaction (PCR) analysis revealed possible TATA and GGA motifs upstream of the 2-CysPrx gene that were recognized by nuclear-encoded plastid RNA polymerase (NEP) as well as a possible −10 and −35 box recognized by plastid-encoded plastid RNA polymerase (PEP). These results suggested the possibility that the C. marina 2-CysPrx gene is found in the chloroplast, and its transcription can be regulated by both NEP and PEP [51].
A previous study [53] reported that the chloroplast-encoding gene is transcribed by NEP or PEP. In general, PEP involves photosynthesis-related genes, and NEP involves housekeeping genes. The authors mentioned three types of NEP (Ia, Ib, and II). Type Ib NEP recognized the sequence YRTA (TATA) motif near the transcription–initiation site and a conserved GGA motif about 18–20 bp upstream of the YRTA motif. The YRTA motif was also reported by Joshi [54] to be located 32 ± 7 bp upstream of the transcription–initiation site. In the case of C. marina, the TATA motif was found 27 bp upstream of the transcription–initiation site, and the GGA motif was found 18 bp upstream of the TATA motif. In contrast, Börner et al. [53] noted that PEP recognizes bacterial σ70 promoters of the −10 (TATAAT) and −35 box (TTGACA). We observed the sequences TAAAAT around 10 bp and TTGATC around 35 bp upstream of the transcription–initiation site in the C. marina 2-CysPrx gene. Those sequences resembled the −10 and −35 box in the bacterial σ70 promoters. The C. marina 2-CysPrx gene thus had similar characteristics of both the NEP and PEP promoters in its transcription–regulation site. Magee and Kavanagh [55] reported that transcriptional patterns in plastid genes and operons can be assigned to three classes, those that contain (1) PEP promoters only, (2) both PEP and NEP promoters, and (3) NEP promoters only. There is hence a possibility that the C. marina 2-CysPrx gene is transcribed by NEP and PEP promoters, although the NEP gene (rpoT;3) remains undiscovered in marine algae [56]. Further analysis will be needed to reveal the mechanisms that regulate the 2-CysPrx gene in C. marina.
This entry is adapted from the peer-reviewed paper 10.3390/antiox10020162
References
- Kim, K.; Kim, I.H.; Lee, K.Y.; Rhee, S.G.; Stadtman, E.R. The isolation and purification of a specific “protector” protein which inhibits enzyme inactivation by a thiol/Fe(III)/O2 mixed-function oxidation system. J. Biol. Chem. 1988, 263, 4704–4711.
- Rhee, S.G.; Kil, I.S. Multiple functions and regulation of mammalian peroxiredoxins. Annu. Rev. Biochem. 2017, 86, 749–775.
- Yamamoto, H.; Miyake, C.; Dietz, K.J.; Tomizawa, K.; Murata, N.; Yokota, A. Thioredoxin peroxidase in the cyanobacterium Synechocystis sp. PCC6803. FEBS Lett. 1999, 477, 269–273.
- Dietz, K.J. Peroxiredoxins in plants and cyanobacteria. Antioxid. Redox Signal. 2011, 15, 1129–1159.
- Fujii, J.; Ikeda, Y. Advances in our understanding of peroxiredoxin, a multifunctional, mammalian redox protein. Redox Rep. 2002, 7, 123–130.
- Nott, A.; Jung, H.S.; Koussevitzky, S.; Chory, J. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 2006, 57, 739–759.
- Dayer, R.; Fischert, B.B.; Eggen, R.I.L.; Lemaire, S.D. The peroxiredoxin and glutathione peroxidase families in Chlamydomonas reinhardtii. Genetics 2008, 179, 41–57.
- Tomanek, K. Proteomic responses to environmentally induced oxidative stress. J. Exp. Biol. 2015, 218, 1867–1879.
- Dietz, K.J. Thiol-based peroxidases and ascorbate peroxidases: Why plants rely on multiple peroxidase systems in the photosynthesizing chloroplast? Mol. Cells 2016, 39, 20–25.
- Okaichi, T.; Yanagi, T. Sustainable development in the Seto Inland Sea, Japan. Terra Sci. Publ. Co. (Terrapub) 1997, 251–304.
- Onitsuka, G.; Aoki, K.; Matsuyama, Y.; Kimoto, K.; Matsuo, H.; Kitadai, Y.; Nishi, H.; Tahara, Y.; Sakurada, K. Short-term dynamics of a Chattonella antiqua bloom in the Yatsushiro Sea, Japan, in summer 2010: Characteristics of its appearance in the southern area. Bull. Jpn. Soc. Fish. Oceanogr. 2011, 75, 143–153.
- Hallegraeff, G.M.; Munday, B.L.; Baden, D.G.; Whitney, P.L. Chattonella marina Raphidophyte bloom associated with mortality of cultured bluefin tuna (Thunnus maccoyii) in South Australia. In Harmful Algae; Reguera, B., Blanco, J., Fernandez, M.L., Wyatt, T., Eds.; Xunta de Galicia, IOC of UNESCO, Grafisant: Santiago de Compostela, Spain, 1998; pp. 93–96.
- Elbraechter, M. Exotic flagellates of coastal North Sea waters. Helgoländer Wiss. Meeresunters 1999, 52, 235–242.
- Amano, K.; Watanabe, M.; Kohata, K.; Harada, S. Conditions necessary for Chattonella antiqua red tide outbreaks. Limnol. Oceanogr. 1998, 43, 117–128.
- Marshall, J.A.; Hallegraeff, G.M. Comparative ecophysiology of the harmful alga Chattonella marina (Raphidophyceae) from South Australian and Japanese waters. J. Plankton Res. 1999, 21, 1809–1822.
- Katano, T.; Yoshida, M.; Yamaguchi, S.; Yoshino, K.; Hamada, T.; Koriyama, M.; Hayami, Y. Effect of nutrient concentration and salinity on diel vertical migration of Chattonella marina (Raphidophyceae). Mar. Biol. Res. 2014, 10, 1007–1018.
- Satta, C.T.; Padedda, B.M.; Sechi, N.; Pulina, S.; Loria, A.; Lugliè, A. Multiannual Chattonella subsalsa Biecheler (Raphidophyceae) blooms in a Mediterranean lagoon (Santa Giusta Lagoon, Sardinia Island, Italy). Harmful Algae 2017, 67, 61–73.
- Giddings, S.N.; MacCready, P.; Hickey, B.M.; Banas, N.S.; Davis, K.A.; Siedlecki, S.A.; Trainer, V.L.; Kudela, R.M.; Pelland, N.A.; Connolly, T.P. Hindcasts of potential harmful algal bloom transport pathways on the Pacific Northwest coast. J. Geophys. Res. 2014, 4, 2439–2461.
- Pinto, L.; Mateus, M.; Silva, A. Modeling the transport pathways of harmful algal blooms in the Iberian coast. Harmful Algae 2016, 53, 8–16.
- Yamaguchi, M.; Imai, I.; Honjo, T. Effects of temperature, salinity and irradiance on the growth rates of the noxious red tide flagellates Chattonella antiqua and C. marina (Raphidophyceae). Nippon Suisan Gakkaishi 1991, 57, 1277–1284.
- Qiu, X.; Shimasaki, Y.; Tsuyama, M.; Yamada, T.; Kuwahara, R.; Kawaguchi, M.; Honda, M.; Gunjikake, H.; Tasmin, R.; Shimizu, M.; et al. Growth phase dependent variation of photosynthetic activity and cellular protein expression profile in harmful raphidophyte Chattonella antiqua. Biosci. Biotechnol. Biochem. 2013, 77, 46–52.
- Portune, K.J.; Cary, S.C.; Warner, M.E. Antioxidant enzyme response and reactive oxygen species production in marine raphydophytes. J. Phycol. 2010, 46, 1161–1171.
- Miao, Y.L.D.; Wang, P.; Wang, X.; Chen, J.; Miao, C.; Song, C. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 2006, 18, 2749–2766.
- Machida, T.; Ishibashi, A.; Kirino, A.; Sato, J.; Kawasaki, S.; Niimura, Y.; Honjoh, K.; Miyamoto, T. Chloroplast NADPH-dependent thioredoxin reductase from Chlorella vulgaris alleviates environmental stresses in yeast together with 2-Cys Peroxiredoxin. PLoS ONE 2012, 7, 45988.
- Ribeiro, C.W.; Korbes, A.P.; Garighan, J.A.; Jardim-Messeder, D.; Carvalho, F.E.L.; Sousa, R.H.V.; Caverzan, A.; Teixeira, F.K.; Silveira, J.A.G.; Margis-Pinheiro, M. Rice peroxisomal ascorbate peroxidase knockdown affects ROS signaling and triggers early leaf senescence. Plant Sci. 2017, 263, 55–65.
- Oda, T.; Nakamura, A.; Shikayama, M.; Kawano, I.; Ishimatsu, A.; Muramatsu, T. Generation of reactive oxygen species by raphidophycean phytoplankton. Biosci. Biotechnol. Biochem. 1997, 61, 1658–1662.
- Oda, T.; Moritomi, J.; Kawano, I.; Hamaguchi, S.; Ishimatsu, A.; Muramatsu, T. Catalase- and superoxide dismutase-induced morphological changes and growth inhibition in the red tide phytoplankton Chattonella marina. Biosci. Biotechnol. Biochem. 1995, 59, 2044–2048.
- Johnson, K.J.; Fantone, J.C.; Kaplan, P.A. In vivo damage of rat lungs by oxygen metabolites. J. Clin. Investig. 1981, 67, 983–993.
- Halliwell, B.; Gutteridge, J.M. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 1984, 219, 1–14.
- Yamamoto, S. Enzymatic lipid-peroxidation-reactions of mammalian lipoxygenases. Free Radic. Biol. Med. 1991, 10, 149–159.
- Yin, H.; Xu, L.; Porter, N.A. Free radical lipid peroxidation: Mechanisms and analysis. Chem. Rev. 2011, 111, 5944–5972.
- Ayala, A.; Muñoz, F.M.; Argüelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell Longev. 2014, 2014, 360438.
- Chan, L.L.; Hodgkiss, I.J.; Wan, J.M.F.; Lum, J.H.K.; Mak, A.S.C.; Sit, W.H.; Lo, S.C.L. Proteomic study of a model causative agent of harmful algal blooms, Prorocentrum triestinum II: The use of differentially expressed protein profiles under different growth phases and growth conditions for bloom prediction. Proteomics 2004, 4, 3214–3226.
- Zhang, S.; Sui, Z.; Chang, L.; Kang, K.; Ma, J.; Kong, F.; Zhou, W.; Wang, J.; Guo, L.; Geng, H.; et al. Transcriptome de novo assembly sequencing and analysis of the toxic dinoflagellate Alexandrium catenella using the Illumina platform. Gene 2014, 537, 285–293.
- Kimura, K.; Okuda, S.; Nakayama, K.; Shikata, T.; Takahashi, F.; Yamaguchi, H.; Skamoto, S.; Yamaguchi, M.; Tomaru, Y. RNA Sequencing revealed numerous polyketide synthase genes in the harmful dinoflagellate Karenia mikimotoi. PLoS ONE 2015, 10, 0142731.
- Shikata, T.; Takahashi, F.; Nishide, H.; Shigenobu, S.; Kamei, Y.; Sakamoto, S.; Yuasa, K.; Nishiyama, Y.; Yamasaki, Y.; Uchiyama, I. RNA-Seq Analysis reveals genes related to photoreception, nutrient uptake, and toxicity in a noxious red-tide raphidophyte Chattonella antiqua. Front. Microbiol. 2019, 10, 1764.
- Hano, T.; Tomaru, Y. Metabolomics-based approach to explore growth phase-dependent markers lit in cultured diatom Chaetoceros tenuissimus. J. Chromatogr. B 2019, 1128, 121779.
- Vingiani, G.M.; Štālberga, D.; De Luca, P.; Ianora, A.; De Luca, D.; Lauritano, C. De novo transcriptome of the non-saxitoxin producing Alexandrium tamutum reveals new insights on harmful dinoflagellates. Mar. Drugs 2020, 18, 386.
- Chae, H.Z.; Kim, H.J.; Kang, S.W.; Rhee, S.G. Characterization of three isoforms of mammalian peroxiredoxin that reduce peroxides in the presence of thioredoxin. Diabetes Res. Clin. Pract. 1999, 45, 101–112.
- Qiu, X.; Mukai, K.; Shimasaki, Y.; Tsuyama, M.; Matsubara, T.; Nakajima, Y.; Honjo, T.; Oshima, Y. Potential maximum quantum yield of photosystem II reflects the growth rate of Chattonella marina in field bloom samples. J. Fac. Agric. Kyushu Univ. 2016, 61, 331–335.
- Qiu, X.; Mukai, K.; Shimasaki, Y.; Tsuyama, M.; Matsubara, T.; Nakashima, T.; Ichinose, H.; Nakazima, Y.; Honjo, T.; Oshima, Y. Variations in the expression of photosynthesis–related proteins in field Chattonella marina cells during a harmful algal bloom. J. Fac. Agric. Kyushu Univ. 2017, 62, 373–380.
- Dietz, K.J.; Horling, F.; Konig, J.; Baier, M. The function of the chloroplast 2-cysteine peroxiredoxin in peroxide detoxification and its regulation. J. Exp. Bot. 2002, 53, 1321–1329.
- Baier, M.; Dietz, K.J. Primary structure and expression of plant homologues of animal and fungal thioredoxin-dependent peroxide reductases and bacterial alkyl hydroperoxide reductases. Plant Mol. Biol. 1996, 31, 553–564.
- Baier, M.; Dietz, K.J. Protective function of chloroplast 2-cysteine peroxiredoxin in photosynthesis. Evidence from transgenic Arabidopsis. Plant Physiol. 1999, 119, 1407–1414.
- Horling, F.; Baier, M.; Dietz, K.J. Redox-regulation of the expression of the peroxide-detoxifying chloroplast 2-Cys peroxiredoxin in the liverwort Riccia fluitans. Planta 2001, 214, 304–313.
- Klughammer, B.; Baier, M.; Dietz, K.J. Inactivation by gene disruption of 2-cysteine-peroxiredoxin in Synechocystis sp. PCC 6803 leads to increased stress sensitivity. Physiol. Plant. 1998, 104, 699–706.
- Sandrini, G.; Piel, T.; Xu, T.S.; White, E.; Qin, H.J.; Slot, P.C.; Huisman, J.; Visser, P.M. Sensitivity to hydrogen peroxide of the bloom-forming cyanobacterium Microcystis PCC 7806 depends on nutrient availability. Harmful Algae 2020, 99, 101916.
- Kim, D.; Watanabe, M.; Nakayasu, Y.; Kohata, K. Production of superoxide anion and hydrogen peroxide associated with cell growth of Chattonella antiqua. Aquat. Microb. Ecol. 2004, 35, 57–64.
- Yuasa, K.; Shikata, T.; Ichikawa, T.; Tamura, Y.; Nishiyama, Y. Nutrient deficiency stimulates the production of superoxide in the noxious red-tide-forming raphidophyte Chattonella antiqua. Harmful Algae 2020, 99, 101938.
- Zachary, Z.A.; Poole, L.B.; Karplus, P.A. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 2003, 300, 650–653.
- Mukai, K.; Teramoto, A.; Qiu, X.; Shimasaki, Y.; Kato-Unoki, Y.; Lee, J.M.; Mizoguchi, M.; Khanam, M.R.M.; Satone, H.; Tatsuke, T.; et al. Gene structure and cDNA sequence of 2-Cys peroxiredoxin in the harmful algal bloom species Chattonella marina and its gene transcription under different light intensities. Eur. J. Phycol. 2018, 53, 29–38.
- Cattolico, R.A.; Jacobs, M.A.; Zhou, Y.; Chang, J.; Duplessis, M.; Lybrand, T.; Mckay, J.; Ong, H.C.; Sims, E.; Rocap, G. Chloroplast genome sequencing analysis of Heterosigma akashiwo CCMP452 (West Atlantic) and NIES293 (West Pacific) strains. BMC Genomics 2008, 9, 211.
- Börner, T.; Aleynikova, A.Y.; Zubo, Y.O.; Kusnetsov, V.V. Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim. Biophys. Acta Bioenerg. 2015, 9, 761–769.
- Joshi, C.P. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987, 15, 6643–6653.
- Magee, A.M.; Kavanagh, T.A. Plastid genes transcribed by the nucleus-encoded plastid RNA polymerase show increased transcript accumulation in transgenic plants expressing a chloroplast-localized phage T7 RNA polymerase. J. Exp. Bot. 2002, 53, 2341–2349.
- Smith, A.C.; Purton, S. The transcriptional apparatus of algal plastids. Eur. J. Phycol. 2002, 37, 301–311.
